Amino Acids and their Properties
advertisement

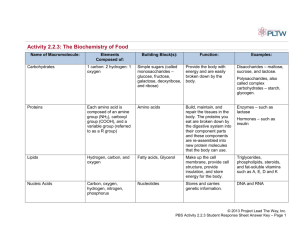
AMINO ACIDS The building blocks for proteins Garrett and Grisham, Biochemistry, Third Edition What You Need to Know • Four basic classes of amino acids – What are general characteristics and importance for each class? – acid-base properties? – How do they form peptide bonds? – What other way can they be classified? • Biological characteristics of some rare amino acids? – i.e. Hydroxylysine, hydroxyproline, gamma-aminobutyric acid (GABA), histamine • What are the acid base properties of amino acids? • What are the spectroscopic properties of amino acids? • Understand the fundamental structural pattern of amino acids in proteins. • Understand protein structure nomenclature. Garrett and Grisham, Biochemistry, Third Edition AMINO ACIDS USED IN LIVIING ORGANISMS GENERAL CONSIDERATIONS • There are a host of amino acids yet only 20 unique amino acids are used to make proteins • Amino acids in proteins contain a central tetrahedral carbon atom • Amino acids polymerize in fabricating proteins via peptide bonds Garrett and Grisham, Biochemistry, Third Edition Amino Acids Building Blocks of Proteins Anatomy of an amino acid. Except for proline and its derivatives, all of the amino acids commonly found in proteins possess this type of structure. Amino Acids Can Join Via Peptide Bonds The -COOH and NH3+ groups of two amino acids can react with the resulting loss of a water molecule to form a covalent amide bond. What is the Fundamental Structural Pattern in Proteins? Peptide formation is the creation of an amide bond between the carboxyl group of one amino acid and the amino group of another amino acid. Garrett and Grisham, Biochemistry, Third Edition The Peptide Bond The trans conformation of the peptide bond. Garrett and Grisham, Biochemistry, Third Edition What is the Fundamental Structural Pattern in Proteins? (a) The peptide bond has partial double bond character. One of the postulated resonance forms is shown here. Garrett and Grisham, Biochemistry, Third Edition What is the Fundamental Structural Pattern in Proteins? (b) The peptide bond has partial double bond character. One of the postulated resonance forms is shown here. Garrett and Grisham, Biochemistry, Third Edition What is the Fundamental Structural Pattern in Proteins? (c) The peptide bond is best described as a resonance hybrid of the forms shown on the two previous slides. Garrett and Grisham, Biochemistry, Third Edition The Peptide Bond • Is usually found in the trans conformation • Has partial (40%) double bond character • Is about 0.133 nm long - shorter than a typical single bond but longer than a double bond • Due to the double bond character, the six atoms of the peptide bond group are always planar • N partially positive; O partially negative Garrett and Grisham, Biochemistry, Third Edition What is the Fundamental Structural Pattern in Proteins? • Proteins are unbranched polymers of amino acids • Amino acids join head-to-tail through formation of covalent peptide bonds • Peptide bond formation results in release of water • The peptide backbone of a protein consists of the repeated sequence –N-Cα-Co• “N” is the amide nitrogen of the amino acid • “Cα” is the alpha-C of the amino acid • “Co” is the carbonyl carbon of the amino acid Garrett and Grisham, Biochemistry, Third Edition What is the Fundamental Structural Pattern in Proteins? The coplanar relationship of the atoms in the amide group is highlighted here by an imaginary shaded plane lying between adjacent α-carbons. Garrett and Grisham, Biochemistry, Third Edition 20 Amino Acids IN PROTEINS On the basis of their R groups the amino acids can be categorized as: Non-polar amino acids Polar, uncharged amino acids Polar, charged amino acids Garrett and Grisham, Biochemistry, Third Edition Note + charge on histidine which has a side chain pKa = 6.4 Thus, 10% of His side chains are positively charged at pH 7.4. Polar, acidic (negatively charged) amino acids Polar, basic (positively charged) amino acids AMINO ACID R GROUPS SUMMARY OF FUNCTIONS • Non polar a) establish folding pattern (globular) b) promote hydrophobic interactions • Polar, uncharged and charged a) establish folding pattern (fibrous) b) promote specific interactions His, Ser, Lys, Asn, Cys Several Amino Acids Occur in Proteins as a result of posttranslational modifications • • • • Hydroxylysine, hydroxyproline - collagen Carboxyglutamate - blood-clotting proteins Pyroglutamate – in bacteriorhodopsin GABA, Histamine, serotonin Garrett and Grisham, Biochemistry, Third Edition Marfan’s Syndrome • Genetic disorder (dominant trait) • Fibrillin protein is mutated • Secondary to mutated fibrillin is excessive transforming growth factor beta (TGF-β) • Weakens tissue causing features of Marfans (long limbs, aortic root dialation, depressed sternum, hyper-flexible joints •Narrow jaws and high arched palate creating dental and orthodontal problems • partially dislocated lens (80% of patients), early onset glaucoma Garrett and Grisham, Biochemistry, Third Edition Garrett and Grisham, Biochemistry, Third Edition Garrett and Grisham, Biochemistry, Third Edition Garrett and Grisham, Biochemistry, Third Edition Titration of glutamic acid. Acid-Base Properties of Amino Acids • Amino Acids are Weak Polyprotic Acids • Each amino acid has at least two chemical groups capable of acting as an acid or base. • Details of these transformations are shown on the next slides. Garrett and Grisham, Biochemistry, Third Edition pKa Values of the Amino Acids You should know these numbers and know what they mean! • Alpha carboxyl group - pKa = 2 • Alpha amino group - pKa = 9 • These numbers are approximate, but entirely suitable for our purposes. Garrett and Grisham, Biochemistry, Third Edition The ionic forms of an amino acid, shown without consideration of any ionizations on the side chain. pKa Values of the Amino Acid R Groups • Arginine, Arg, R: pKa(guanidino group) = 12.5 • Aspartic Acid, Asp, D: pKa = 3.9 • Cysteine, Cys, C: pKa = 8.3 • Glutamic Acid, Glu, E: pKa = 4.3 • Histidine, His, H: pKa = 6.0 Garrett and Grisham, Biochemistry, Third Edition pKa Values of the Amino Acid R Groups, cont. • • • • Lysine, Lys, K: pKa = 10.5 Serine, Ser, S: pKa = 13 Threonine, Thr, T: pKa = 13 Tyrosine, Tyr, Y: pKa = 10.1 Garrett and Grisham, Biochemistry, Third Edition Titration of glycine, a simple amino acid. The isoelectric point, pI, the pH where the molecule has a net charge of 0, is defined as (pK1+ pK2)/2. Titration of lysine. Reactions of Amino Acids • Carboxyl groups form amides & esters • Amino groups form Schiff bases and amides • Side chains show unique reactivities – Cys residues can form disulfides and can be easily alkylated – Few reactions are specific to a single kind of side chain Garrett and Grisham, Biochemistry, Third Edition Spectroscopic Properties • Phe, Tyr, and Trp absorb at UV wavelengths • Absorbance at 280 nm is a good diagnostic device for amino acids • Absorbance at 190 nm is characteristic of peptide bonds and is therefore a good method for detecting proteins which contain few aromatic amino acids. Garrett and Grisham, Biochemistry, Third Edition The ultraviolet absorption spectra of the aromatic amino acids at pH 6. (From Wetlaufer, D.B., 1962. Ultraviolet spectra of proteins and amino acids. Advances in Protein Chemistry 17:303– 390.) Amino Acid Purification Methods • Chromatographic Methods – Ion exchange – Reversed Phase Garrett and Grisham, Biochemistry, Third Edition Cation (a) and anion (b) exchange resins commonly used for biochemical separations. Operation of a cation exchange column, separating a mixture of Asp, Ser, and Lys. a) The cation exchange resin in the beginning, Na+ form. (b) A mixture of Asp, Ser, and Lys is added to the column containing the resin. (c) A gradient of the eluting salt (e.g., NaCl) is added to the column. Asp, the least positively charged amino acid, is eluted first. (d) As the salt concentration increases, Ser is eluted. (e) As the salt concentration is increased further, Lys, the most positively charged of the three amino acids, is eluted last. Chromatographic fractionation of a synthetic mixture of amino acids on ion exchange columns A second column with different buffer conditions is used to resolve the basic amino acids. Reversed phase chromatography of amino acids Garrett and Grisham, Biochemistry, Third Edition The Sequence of Amino Acids in a Protein • • • • Is a unique characteristic of every protein Is encoded by the nucleotide sequence of DNA Is thus a form of genetic information Is read from the amino terminus to the carboxyl terminus Garrett and Grisham, Biochemistry, Third Edition Amino acid composition: Frequencies of the various amino acids in proteins for all the proteins in the SWISS-PROT protein knowledgebase. These data are derived from the amino acid composition of more than 100,000 different proteins (representing more than 40,000,000 amino acid residues). The range is from leucine at 9.55% to tryptophan at 1.18% of all residues. Garrett and Grisham, Biochemistry, Third Edition Proteins - Large and Small Garrett and Grisham, Biochemistry, Third Edition Protein Structure (nomenclature) Peptide=short polymers of amino acids NOTE: each unit=amino acid residue (dipeptide, tripeptide, tetrapeptide) If >12 residues=oligopeptide If several dozen residues=polypeptide Proteins are composed of 1 or more polypeptide chains 1=monomeric proteins 2 or more = multimeric proteins (homomultimeric or heteromultimeric) Multimeric proteins are usually designated by greek letters and subscripts i.e. 2=homodimer 2b2=heteromultimer (tetramer) (4 polypeptides of 2 kinds) Garrett and Grisham, Biochemistry, Third Edition Protein Structure Protein Shape: 2 general classes globular fibrous Primary Structure: amino acid sequence (NH2 to COO end) Secondary Structure: characteristic patterns of the polypeptide chain extending along 1 dimension - caused by hydrogen bonding interactions between adjacent amino acid residues, i.e. helical or pleated sheets Garrett and Grisham, Biochemistry, Third Edition Protein Structure Tertiary Structure: when the polypeptide chains bend and fold to assume a more compact three-dimensional shape Quaternary Structure: subunit organization in proteins Conformation Possibilities: rotational possibilities create many possible orientations for the protein chain proteins have many biological functions, thus they can be classified by their biological roles Garrett and Grisham, Biochemistry, Third Edition Proteins - Large and Small • Insulin - A chain of 21 residues, B chain of 30 residues -total mol. wt. of 5,733 • Glutamine synthetase - 12 subunits of 468 residues each - total mol. wt. of 600,000 • Connectin proteins - alpha - MW 2.8 million • beta connectin - MW of 2.1 million, with a length of 1000 nm -it can stretch to 3000 nm Garrett and Grisham, Biochemistry, Third Edition Proteins - Large and Small Garrett and Grisham, Biochemistry, Third Edition Proteins - Large and Small From Table 4.2 Size of protein molecules. Molecular weights: Insulin, 5,733; Cytochrome c, 12,500; Ribonuclease, 12,640; Lysozyme, 13,930; Myoglobin, 16,980. Garrett and Grisham, Biochemistry, Third Edition Proteins - Large and Small From: Table 4.2 Size of Protein Molecules Molecular weights: Hemoglobin, 64,500; Immunoglobulin, 149,900; Glutamine synthetase, 600,000. Garrett and Grisham, Biochemistry, Third Edition