Review of Lecture 1
advertisement

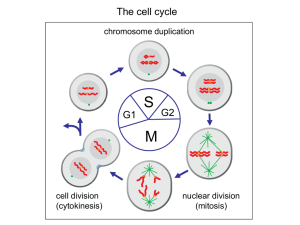
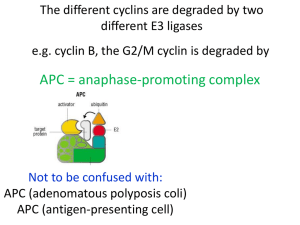
Gene Nomenclature Budding Yeast: CDC28 = wild type gene cdc28 = recessive mutant allele Cdc28 or Cdc28p = CDC28 protein Fission Yeast: cdc2 = the wild type gene cdc2- = recessive mutant allele p34cdc2 = the protein Non-genetic systems: not standard use either yeast, or use unique name with yeast name superscript or whatever Regulation of G1 to S Role in G1 for cdc2/cdc28 at START But cdc2/cdc28 regulators acted at G2 to M ONLY cdc2D cdc13/B-type Cyclins cdc25 and wee1 CKIs wee1 ????? G1????? Cyclins cdc25 cdc2 G2 G1 Mitosis S phase Cyclins Nomenclature All contain “cyclin box” domain Cyclin A Cyclin B’s Vertebrate Cyclin B1, B2 S. pombe cdc13... S. cerevisiae CLB1,2,3,4,5,6 cdc2 binding domain A and B’s closely related -destruction box - required for S and M Differ in expression pattern - “A” accumulates earlier, - degraded earlier G1 Cyclins G1’s more similar to each other S. cerevisiae CLN1, 2, 3 - PEST domains Mammalian Cyclin D, E - required for exit from G1 What do Cyclins do? 1. Positive Regulator of CDK catalytic subunit 2. Potentially impart CDK substrate specificity 3. Potentially impart CKD inhibitor specificity 4. Potentially determine specific CDK localization Cyclins can be grouped by expression patterns - CLN1 and CLN2 and CLB5 and CLB6 - CLB3 and CLB4 - CLB 1 and CLB2 CLN1,2,CLB5,6 CLB1,2 CLB3,4 CLN3 G1 Start Metaphase Yeast Cyclin Genes: CLNs and CLBs Figure 13-22 Quadruple clb1,2,3,4 Mutant S phase Cyclins CLB5, CLB6 NO PHENOTYPE - double mutants -gain of function -???????????? G1 Cyclins: CLN1 CLN2 CLN3 Triple cln1,2,3 mutant G2 Cyclins CLB1 CLB2 CLB3 CLB4 Dominant CLN Mutations Role of Cyclins in G1 to S???? Cdc28/Cln3 “Start-specific” Transcription Cdc28/Cln1,2 Budding SPB dup Sic1 Cdc28/Clb5,6 DNA Rep Regulation of G1 to S Starting point of this study: - Specific role of CLNs in regulation of G1 to S not clear - triple mutant doesn’t bud, dup SPB, or rep DNA - CDC34, CDC53, and CDC4 Required specifically for DNA replication - high Cdc28/Cln activity - Cdc34 = UBC - CLB1,2,3 and 4 not required for S Phase - What about CLB5 and CLB6, and what’s this to do with CDC34 et al ? Clb5/Cdc28 kinase accumulates near/after Start Protein Levels Clb/Cdc28 Activity by Histone H1 Kinase Assay Clb5,6/Cdc28 Necessary and Sufficient for DNA Replication clb1,clb3,clb4,clb5,clb6 clb2(ts) GAL::CLB5 Grows at 23 deg on Galactose medium Dies at 37 degrees on Glucose medium Cdc34 involved in degradation of a Cdc28/Clb inhibitor?? CLBs redundant for S Phase, with 5 and 6 most important Inhibition of Clb/Cdc28 by Sic1 in vitro Cdc28/Clb specific inhibitor accumulates in cdc34 et al mutants Inhibitor depends on SIC1 function Sic1 is a Cdc28 inhibitor In vitro demonstration of Clbspecific inhibition and Sic1/Cdc28/Clb complex formation Implications and Predictions... Clb5,6 expressed in late G1 Clb5,6 major role in S phase induction after Start Clb5,6/Cdc28 and other Clb/Cdc28s held inactive by Sic1 Sic1 function is opposed by cdc34,4,53 after Start… Sic1 should be regulated - on before start, off after start Sic1 regulation dependent on cdc34,4,53 Phenotype of cdc34 mutants should be dependent on Sic1 sic1 mutants should be sensitive to CLB5 over expression Cdc34 dependent Regulation of Sic1 Accumulation Sic1 phosphorylated by Cdc28/Cln in vitro Sic1 destroyed in a CDC34dependent manner Phosphorylation correlates with time of destruction Sic1 is required to prevent S phase in cdc34 G2/M arrest? Regulation of G1/S Cdc28/Cln3 cln1,cln2,cln3 dead cln1,cln2,cln3,sic1 alive “Start-specific” Transcription only essential function of CLNs! Cdc28/Cln1,2 Budding SPB dup Sic1 Cdc28/Clb5,6 DNA Rep Mammalian regulation of G1 to S PICK 1 CDK/CKI Pair Compare and Contrast Roles of G1 CDKs and CKIs in G1 to S in budding yeast and mammalian cells Ubiquitin Dependent Proteolysis in Cell Cycle Regulation Cell Free Cell Cycling Extracts Visualizing Mitotic Spindle Formation and Chromosome Segregation in vitro Cyclin is destroyed at the time of sister chromatid separation by ubiquitin-dependent proteolysis Ubiquitin-dependent proteolysis of other mitotic proteins control separation of sister chromatids Identification of the Anaphase Promoting Complex (aka Cyclosome or APC/C) Cell Free Cell Cycling Extracts Cyclin mRNA is necessary and sufficient to drive cell oscillations 90 = nondegradable NH2-terminus or D-box sufficient for mitotic-specific, ubiquitin-dependent, proteolysis Role of Cyclin B destruction in progression through Mitosis Cdc2/Cyclin B (MPF) activity peaks at metaphase Cyclin destruction correlates with Cdc2/Cyclin B inactivation Regulation of metaphase to anaphase by Cyclin B Proteoysis half right... “Cell Free Mitosis” Reversible Arrest at Metaphase Visualize MTs and DNA Synchronous release from Metaphase-Anaphase-G1 + EGTA (minus calcium) extract arrests in “mitosis” = “CSF extract analogous to mature egg haploid genome + sperm nuclei, + rhodamine-labelled tubulin + calcium, enter G1, S -sister chromatids + “CSF extract”, enter and arrest at metaphase + initiate anaphase by adding calcium again Metaphase to Anaphase in vitro Extract dependent on exogenous Cyclin B mRNA - WT Cyclin B TIME AFTER SECOND CALCIUM ADDITION Extract dependent on exogenous Cyclin B mRNA = 90 Cyclin B TIME AFTER SECOND CALCIUM ADDITION Extract contains endogenous Cyclin B mRNA PLUS D-box peptide Interpretation... Data: Preventing Cyclin B destruction blocks after anaphase Adding competitive inhibitor of Cyclin B destruction blocks at metaphase Interpretation... Cell Cycle Regulation of Ubiquitin Dependent Proteolysis SCF - Skp1, Cdc53, F-box (Cdc4) - Sic1, Cln’s... APC/C - Anaphase Promoting Complex or Cyclosome - Securing (anaphase inhibitor), mitotic Cyclins,... Exam 1 - will write in after Tuesday’s lecture questions on cell cycle analysis using mutants, synchronous cultures - interpret data - design experiment your comparison contrast of G1 to S regulation by Cdk/Cyclin/CKI in mammalian cells and budding yeast review article is sufficient for mammalian cdk info no wrong answers I don’t care how long/short - just thoughtful can bring the answer to class with you questions on cell cycle regulated, ubiquitin-dependent proteolysis - interpret data from in vivo (genetic) and in vitro experiments