Unit 2 - E
advertisement

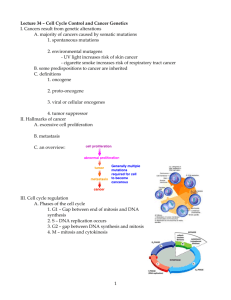
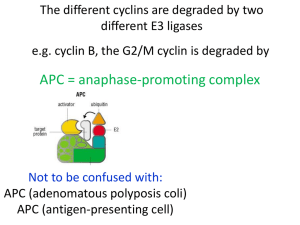
Dr Forsburg's all-purpose Cell Cycle Lecture Notes These notes were used for my lectures in both BIMM112 (UCSD Division of Biology) and BMS210 (UCSD School of Medicine, Biomedical Sciences Program). The 2001 Nobel Prize in Physiology or Medicine was awarded to Lee Hartwell, Paul Nurse, and Tim Hunt for their ground-breaking work on cell cycle regulation. Starting in the late 60s, Hartwell used budding yeast to identify mutants that blocked specific stages of cell cycle progression. Nurse, working in fission yeast in the 70s, went on to isolate mutants that could also speed up the cell cycle, thus focussing his attention on the original CDK kinase, cdc2. In the 80s, Hunt identified proteins in sea urchin extracts, the levels of which varied through the cell cycle hence "cyclins". All three have continued to make important advances in cell cycle research including the identification of checkpoints, mechanisms coupling cell morphology to the cell cycle, and identification of additional classes of kinases, cyclins, and inhibitors. For more information about their studies that led to the award, see this BBC brief. You can also visit their web pages: Hartwell, FHCRC Seattle, Nurse (ICRF London), and Hunt, ICRF-Clare Hall. Background on the pombe cell cycle can be found on our site. Problems printing? Try printing with graphics turned off to see the text properly. Additional slides used in lecture are available here. Reading list of some interesting papers that illuminate the principles discussed here. Rao, P. N., Johnson, R. T. (1970) Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature: 225:159-164. A classic paper indicating tht the cell cycle is regulated by trans-acting fctors. Nurse P. A long twentieth century of the cell cycle and beyond. Cell. 2000 100:71-8. Review. Noton E, Diffley JF CDK inactivation is the only essential function of the APC/C and the mitotic exit network proteins for origin resetting during mitosis.Mol Cell 2000 5 :85-95 Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999 400:3742. Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000 287:1824-7. All students: you are responsible for knowing the UCSD Policy on the Integrity of Scholarship. The Cell Cycle | Evidence for Regulation | Genetic Analysis | Cdc2 Regulation | Cyclins | Inhibitors | Destruction | Mitotic exit | Replication | S to M phase | Checkpoints | Meiosis | Links | What the heck are all these gene names? Play the cell cycle game 1) The cell cycle A process of division allowing the duplication of cells a cycle: end products (daughter cells) are the same as starting products (mother cells) G1 - S - G2 - M phase, where G1/S/G2 represent interphase; stages of mitosis are prophase, metaphase, anaphase, and telophase A chromosome cycle of chromosome duplication and segregation; S phase and M phase must be coordinated but non-overlapping Chromosome duplication occurs in S phase. All the DNA must be copied faithfully exactly once. Must occur in a timely fashion: coordination of individual origins of replication to fire once and only once. Duplicated chromosomes must remain attached to allow subsequent cell cycle events: concept of cohesion. Chromosome segregation occurs during M phase. During prophase, DNA chromosomes condense or package to allow more efficient movement. Cell must assemble a mitotic spindle. At metaphase, the chromosomes align upon spindle, attached via their kinetochores to microtubules. Upon attachment and organization of all the chromosomes, they are segregated by releasing cohesion that attaches the sisters together, and reeling in the spindle (anaphase) until they decondense and form new nuclei (telophase). Click here for more info and diagrams of mitotic cells Question of cell cycle regulation: how to ensure the orderly progression of events so that nuclear cycle is coordinated with cell growth and physical separation. Replication must occur once per cell cycle and precede chromosome segregation; segregation must be complete before cytokinesis (cell division) Question of chromosome dynamics: the cell cycle is also a chromosome cycle 2) Evidence for cell cycle regulation Convincing evidence for actual cell cycle regulators came from several key experiments. Rao and Johnston: mixing nuclei together in the same cytoplasm (heterokaryon) to determine whether they could influence one another. o S + G1 : induces the G1 nuclei to start S phase. Suggests that the S phase nucleus contains a diffusable factor that will induce replication. o S + G2 phase: the G2 nucleus does not do S phase. Something about G2 phase is refractory to the diffusible factor from S phase. o G1 + G2 phase: no S or M phase o M phase + interphase: induces inappropriate mitosis o Conclusion: there are diffusible factors that can promote S or M phase. The S phase promoting factor only works on G1 nuclei. The m phase promoter works on everything. Xenopus extracts: Take oocytes blocked before their final division (like M phase), ask is it possible to induce them to enter M phase by injecting them with cytoplasm from post-M phase eggs. Answer: Yes. Purify and find two proteins comprising an activity called MPF. Sea urchin embryoes. Find when looking at total proteins in a population undergoing synchronous division that some proteins go up and down with the cell cycle: cyclin. Yeast cells: it is possible to isolate yeast mutants that can grow (e.g., synthesize macromolecules) but cannot divide their nuclei: deficient in proteins required specifically for cell division. 3) Key work required genetics. Genetic analysis in simple cells provides blueprint for biochemical studies Yeast cells, which are simple, single celled eukaryotes, undergo cell division cycle like human cells. Yeast is a general term, like "animal", so that it covers organisms as diverse and humans and worms. Two types of yeast were used. The key is that yeast cells are (a) simple and (b) haploid, that is one copy of each chromosome. Saccharomyces cerevisiae: budding yeast. o Simple cell that divides by budding. Unusually short G2 phase. Brewer's or Baker's yeast. Popular model for cell biology problems Schizosaccharomyces pombe: fission yeast o pombe means beer in Swahili. More typical cell cycle with long G2. Good model for studies of growth control. For both yeasts: key insight was the idea that it might be possible to identify genes required to regulate cell division by looking for mutants. Key to this: haploid cells. expect genes for cell division to be essential. Look for mutants that are conditional: that is mutant only in one condition, not another. Use temperature. TS (temperature sensitive) mutants are unable to do their job at high temperature, presumably due to altered protein structure. At low temps, proteins stay intact and can grow cells. Isolate cells with mutations, ask what happens at high temp? cells will get "stuck" at a particular cell cycle stage. o How to determine the stage at which blocked? use landmarks. For example, did they replicate their DNA or not? For budding yeast: is there a bud and how big? In fission yeast, cells simply elongate For both: is there a mitotic spindle? Did nuclei condense? cdc mutants ("cell division cycle") o If it is possible to remove a key component, is it also possible to change regulation/timing? Model: if you slow down the fission yeast cell cycle, get long. if you speed it up, get short. isolate short or "wee" mutants: defective in timing or regulation. using methods of genetics, determine how many genes are mutated. That is, for all the mutants that arrest cells in G2, how many different genes do they represent? combine mutants together to see if it is possible to determine pathways between them. 4) isolation of Cdc2 kinase and determining regulation Fission yeast: two different types of mutation in gene called cdc2: elongated mutant (ie, cell cycle delay), and small mutant (ie, cell cycle speeds up). Other phenotypes will be discussed later in lectures Suggests that cdc2 may function in timing: required for division (long mutant) but changing its regulation changes timing (short mutant). Mutants and phenotypes: mutant name: means protein not active. OP: means producing too much of the normal protein: essentially over-active. LOF (loss of function), and GOF (Gain of function), respectively. cdc25 long wee1 short cdc2-L long cdc2-w short OP cdc25 short OPwee1 long cdc13 long Suggests that changing activity of these proteins can change how they behave. How to determine order of events? Combine mutants and ask what the double mutant looks like. Called epistasis. If one protein is required for cell cycle, then changing activity of protein s upstream won't make a difference if protein 1 is missing. Requires the mutants have different phenotypes (appearance) to distinguish which phenotype is displayed. Results: cdc2-L wee1 long cdc2-L OP cdc25 long cdc2-w cdc25 short cdc2-w OP wee1 short cdc25 wee1 normal Conclusion: wee1 and cdc25 act upstream of cdc2. wee1 inhibits cdc2: because absence of wee1 speeds things up. But if cdc2 is missing wee1 doesn't matter. cdc25 activates cdc2, because if cdc25 is missing, things slow. but if cdc2 already active (cdc2-w), cdc25 doesn't matter. cdc13 works with or downstream of cdc2 (can't do experiment reciprocally to be sure). Important: cdc25 and wee1 antagonize each other, because together everything looks okay. These yeast experiments agreed with biochemical data. Showed that cdc2 and cdc13 bind together. Yeast cdc2/cdc13 turn out to be the same two proteins that bound together in Xenopus MPF. Also showed cyclin (Cdc13) levels go up and down as cell goes through cell cycle. cdc2 is a kinase that is only active if bound to cyclin. Found that it is itself phosphorylated on Y15 by wee1, and this phosphate is removed by cdc25. Wee1 is a kinase, cdc25 a phosphatase. Additional biochemical and genetic experiments showed an additional phosphorylation on T161, activating. o cdc2 alone: not active o cdc2 cdc13 : not active o cdc2 (Y15 Phosphorylated) cdc13: not active o cdc2 (Y15 Phos T161Phos): not active o cdc2 (T161Phos) cdc13: active Basic cell cycle regulation is regulation of state of cdc2. requires 2 things: cyclin partner and appropriate phosphorylation. Cyclin accumulates during cell cycle, destroyed during M phase. phosphorylation provides additional switch. Phenotype of cdc2 Y15 mutant: chronically activated. Relies on accumulation of cyclin. Insensitive to wee1/cdc25 "switch". Dangerously active kinase, cannot respond to damage (see below) 5) Cyclin/CDK cycles in the cell cycle Also apparent that other major transitions in cell cycle regulated by cyclin/CDK ("cyclin dependent kinases"). other cyclins that peak earlier -- G1, S phase cyclins. In yeast, only one CDK, multiple cyclins. Presumed that cyclins make kinase specific for particular substrates. Periodic activation of kinase via association with different cyclins. In S. cerevisiae, several overlapping cyclin activities: G1 cyclins CLN1-3, S cyclins CLB5-6, M cyclin CLB1-4 In humans, multiple CDKs as well, so overlapping activation of different complexes. CDK4/cyclin D -> CDK2/cyclin E -> CDK2/cyclin A -> cdc2/Cyclin B How is the transition accomplished, to assure cell cycle moves forward? three broad methods: o 1) Phosphorylation/dephos of CDK (as described) o 2)Specific inhibitors to regulate CDK activity. o 3) Destruction of cyclin and inhibitors at appropriate time in cell cycle 6) Inhibitors of cell cycle CKI, or Cyclin Kinase Inhibitors. prevent accumulated cyclin/CDK from acting. Probably dosage dependent. p21, p27, p16 in human cells. In yeasts, SIC1 or rum1. As yeast cells enter G1, specific inhibitor of B-cyclins is present (SIC1/Rum1). Keeps the mitotic kinase from acting either too long, or too soon. As cells proceed into S phase, destruction of this inhibitor is triggered. In budding yeast, Sic1 active as cells exit mitosis and enter G1. At the same time, a special set of G1 cyclins is transcribed and synthesized. Sic1 only acts on G2 cyclins (B class). The CDK/G1 cyclin phosphorylates SIC1. The phosphorylated Sic1 is then bound by a complex called the SCF, which targets it for destruction (discussed below). Thus, the cycle is set up to (A) prevent premature activation of the NEXT set of cyclins and (B) ensure that Sic1 is turned off as the current set gets activated. In Fission yeast, Rum1 protein works like budding yeast SIC1. Although they do the same job, these proteins are not related to one another by sequence. Other inhibitors are regulated analogously. Direct inhibition of CDK activity is not the only way to block cell cycle progression. For example, the Rb protein in human cells binds to the E2F transcription factor and prevents its activity. no E2F, no S phase genes are expressed. Just as in yeast the increasing levels of the G1 cyclins lead to inactivation of SIC1 protein, so in human cells does increase of CDK4/cycD and then CDK/cycE lead to inactivation of Rb. As CDK activity increases, Rb more phosphorylated. This causes Rb to release E2F transcription factor, which then can turn on S phase genes. Rb is dephosphorylated late in the cell cycle to prevent further division unless appropriate cyclin activity is present. Thus Rb is DOWNSTREAM of CDK activity, while CKIs are UPSTREAM. In mammals, two broad classes of CKI: INK4 family (p15/p16/p19) specific for CDK4/Cyclin D; probably regulates cell cycle entry in response to growth factors etc. CIP/KIP family (p21, p27) induced by p53, may mediate normal control and cell cycle response to damage. Interestingly p21 may act both positively and negatively, depending upon how much is present. CIP/KIPs can regulate all classes of CDK/cyclins, at least in vitro. 7) Regulated destruction A number of proteins are regulated by turnover: proteolysis. Ensures that cell cycle can't roll backward. This requires that the targets be ubiquitinated by specific ubiquitn ligases, which targets them to the proteosome for destruction. o Degradation of cyclin is essential to keep cell cycle moving forward. Making a cyclin mutant that cannot be degraded traps cells in M phase. o CKIs, such as Sic1 and Rum1, must also be turned over. o Cohesion between sister chromatids must be removed: must degrade "molecular glue" APC identified by isolation of mutants that affect protein stability in mitosis. mutants that fail to turn over proteins that must be destroyed for cell cycle progression, will block at that point in the cell cycle. Expression of a "nondegradeable cyclin" has the same phenotype as mutation of the protein that degrades cyclin. In this way, isolate components APC (anaphase promoting complex, or cyclosome) required for ubiquitination of substrates and targeting to proteasome. Two points of destruction: metaphase to anaphase transition, when chromosomes separate, and mitotic exit, when cyclin degraded. These were distinguished because expression of a non-degradeable cyclin did not prevent chromosome segregation o o o o o o o APC works through two sets of targets because it has two specificity factors. First, molecular glue (metaphase to anaphase transition). Protein PDS1 ("securin") binds ESP1 ("separin") and cohesion between chromosomes remains intact. If EPS1 is freed from PDS1, chromosomes will separate. Thus, destruction of PDS1 mediated by APC leads to chromosome separation. If pds1 is mutant, this will happen prematurely. APC targeting to PDS1 is provided by CDC20 subunit. Activation of this APC/CDC20 requires mitotic CDK activity (makes sense since CDK promotes mitosis!) mutants in substrates show that APC independently affects meta->ana transition and cyclin degradation/exit. Target sequence is called "destruction box". CLB2 lacking destruction box blocks after anaphase; PDS1 still degraded. However PDS1 affects cyclin turnover by blocking activation of APC/CDH1: crosstalk between the two APC subunits. PDS1 lacking destruction box inhibits degradation of CLB2 and ASE1. Still Pds1 is not essential for viability in yeast. This system is not sufficient to allow mitotic exit and cyclin turnover. Different specificity factor CDH1/HCT1 required here. Inactivated by CDK; when APC/CDC20 gets activated, that inactivates CLB5 and thus allows APC/CDH1 to be activated sequentially. In turn, this inactivates CLB2, allowing cells to proceed out of mitosis. Additional kinases also involved in pathway to release cells to G1 phase (see mitotic exit, below). Polo-kinase (Cdc5 in budding yeast) is required for activation of APC/CDH1. Note that activation of APC/CDH1 will inactivate the mitotic kinase--which activated APC/CDc20. This ensures forward momentum. net effect: cell cycle can't "roll backward" As ever, system may vary in different cell types. In fission yeast and other eukaryotes, the CDH1-homologue appears to be responsible for degrading the mitotic cyclin in G1 phase, NOT in the exit from mitosis. Thus, the "second" form of the APC may be G1-specific. This may relate to observations in fission yeast that the mitotic exit network - equivalent proteins act in septation....which occurs after mitotic exit. SCF in S phase, and APC are related complexes that recognize specific substrates and target for destruction. Both are types of ubiquitin ligase (E2) enzymes that covalently attach the small peptide ubiquitin to targets. Ubiquitinated proteins are targeted by the proteasome, which degrades them SCF ubiquitinates proteins in G1 to S phase (e.g., SIC1). Contains several subunits including "cullin" (CDC53 in yeast), SKP1 linker protein, and specificity factor containing an "F box sequence" (CDC4 in yeast) that binds to phosphorylated substrate. These proteins are distinct from APC but share some motifs. As CKI (SIC1) is expressed, it inhibits the downstream CLB kinase. But at the same time, the CLN form of the kinase is induced, leading to the phosphorylation of Sic1, and its recognition by the SCF. This results in SIC1 turnover, and release of inhibition of S phase kinase. F box factor itself is ultimately targeted for destruction. Other specificity factors (F box proteins) can association with the cullin-SKP1 base and thus its activity not limited to G1 or even cell cycle regulation. However, its substrates must be phosphorylated, and thus recognized by F box factors. 8) Regulation of mitotic exit: from one cell cycle to another Mitotic exit network (MEN) coordinates APC with inactivation of CDK--one more layer of complication, one more mechanism for irreversability. The network consists of a GTPase-activated kinase cascade that ultimately regulates the activity of a phosphatase, CDC14. In budding yeast an important part of this regulation is spatial: the MEN is activated when one of the spindle poles moves to the new bud. (It is also affected by the spindle checkpoint, discussed below) This leads to the release of CDC14 phosphatase from the nucleolus, where it has been sequestered by NET1 protein. CDC14 phsophatase antagonizes the mitotic CDK by dephosphorylating the same substrates. Thus, whether cell will exit from mitosis will be determined by the relative balance of the kinase and the phosphatase. The MEN is inhibited by the spindle checkpoint, and by the activity of PDS1. CDC14 also dephosphroylates CDH1, leading to increased activity of the APC/CDH1, and further inactivation of CLB2. Because the mitotic form of the kinase prevents formation of the replication complex called the preRC (see below), the action of CDC14 not only helps push the cells out of mitosis but facilitates the next S phase. CDC14 release leads to the activation of SIC1 by two pathways:first, by dephosphorylating the SIC1 transcription factor SWI5, which allows SWI5 to enter the nucleus and activate SIC1 expression, and second, by directly dephosphorylating SIC1, and thus protecting it from degradation. However, as the CLN cyclins accumulate, they will eventually overcome the CDC14 dephosphorylation of SIC1, and this will drive the cycle forward. In fission yeast, the CDC14-equivalent protein is not essential, and the components of the MEN are not involved in promoting mitotic exit. rather, they form a septation-inducing network (SIN). As discussed above, the APC/CDH equivalent complex also appears to be G1, rather than late-M specific. Thus the same sets of proteins can be used to accomplish subtly different requirements. 9) Regulation of replication onset: downstream of CDK Another example of how interplay of CDK, degradation, and CKI work is in the regulation of the onset of DNA replication. Problem: Must ensure that replication origins finally only once per cell cycle, and fire after mitosis is complete. Solution: couple assembly of structures that fire replication to CDK activity. Mitotic CDK activity prevents formation of active origin structures (prereplicative complex, or pre RC. Thus, only when mitotic CDK activity (B cyclin) is low can complex assemble. Thus preRC assembly is limited to late M phase/G1. However, firing of this complex, and activation of replicaiton, requires CDK activity. Thus, CDK activity both activates the origin and prevents it from reactivating, all at once. Components required: o Origin of Replication (yeast ARS: Autonomously Replicating Sequence) on the DNA: site where DNA replication begins. o ORC complex, identifying the origin throughout the cell cycle o Cdc18 (AKA Cdc6) protein: activates origin, allows loading of additional factors. targeted for destruction by mitotic CDK. o MCM proteins: together, form likely helicase. Can only load at origin when CDC18 is there. o replication machinery: polymerases, ligases, etc. Require that MCMs are loaded first. Example from S pombe. Note: put the cursor over the righthand image to see an animated version! o Cdc18 protein is transcribed specifically before S phase, in M/G1 phase. o But CDK activity is high, e.g. in M or S phase, the origin cannot be assembled because Cdc18 is targeted for destruction by CDK phosphorylation o Once origin is assembled, CDK activity required to fire it (exactly how remains unclear) o premature activation is prevented by CKI Rum1p, which keeps CDK activity low o Deletion of Cdc18 results in failure initiate DNA replication. OP-rum1 (like SIC1, a CDK inhibitor) leads to inhibition of kinase, failure to degrade Cdc18, re-initiation of origins. o Independent pathways also involved, including another kinase CDC7 (Hsk1 in the figure below), and sequential loading of different replication factors. o (See this page for complete description) 10) Dependency of mitosis on S phase Cell cycle requires sequence of S phase - M phase. Maintained in part by regulation of CDK. For example, mutations in S. pombe that affect mitotic CDK activity can lead to "re-replication" in which cells repeat S phase without any intervening M phase (see diagram of phenotypes, above). S. pombe is particularly easy to manipulate in this way. Mutations that cause rereplication include : o temperature sensitive mutations of Cdc2 kinase (when returned to permissive temperature), suggesting that the kinase is re-set in some way. o Overproduction of the CKI Rum1 (roughly equal to SIC1 in cerevisiae o Deletion of the mitotic cyclin Cdc13 o Overproduction of Cdc18, an S phase inducer. Cdc18 is a CDK substrate, and when overproduced may saturate the CDK system. Phosphorylated Cdc18 is degraded by the SCF. Mitotic progression is also directly dependent upon S phase. For animation, move your cursor over the image at the right o Cohesion between sister chromatids is established during S phase o Absence of cohesion leads to premature chromosome separation and segregation defects o Normal cohesion is required to establish tension between the chromosomes on the mitotic spindle, and also to orient the chromosomes properly for the mitotic divisions. o Cohesin connections are dissolved by ESP1 (separin) when PDS1 (securin) is destroyed by APC 11) Checkpoints Checkpoints maintain the order of unrelated events by signaling if something goes wrong, e.g. Damage in G1 or G2, incomplete replication, incomplete establishment of mitotic apparatus. Characteristic of checkpoints is that they are usually not essential (pace, Don...). This is particularly true of the checkpoints in yeast As long as everything works all right, then no problem. However, if anything is perturbed--damage occurs, S phase takes too long, spindles fail to assemble, then checkpoint becomes essential to prevent continuation of the cell cycle. In checkpoint mutants, mechanical apparatus of division is intact, and the problem is regulatory. in multi-cellular organisms, some checkpoints are essential for viability of the organism. Generally, even in cell types where some checkpoints are essential, we can think of them as extrinsic to normal engine of the cell cycle--an additional layer of regulation on top of the CDK/cyclin engine. A checkpoint response consist of three broad components: something to generate the signal, something to transduce it, and something to receive it. The transducers are typically what we think of as "checkpoint proteins". how to identify checkpoint proteins, if they aren't essential? Key is to make them essential. For example, in response to irradiation, most yeast cells will arrest the cell cycle, repair the damage, and then continue. A cell that cannot repair the damage will arrest permanently. A cell that can repair the damage but can't arrest will go on to divide, with lethal consequences. Difference is arresting as one cell, or as a microcolony. Similarly, treatment with low levels of a spindle poison such as benomyl will lead to transient arrest and recovery in wild type. Lethally sensitive mutant such as tubulin mutants will die as single cells. Checkpoint mutant cells continue to try to divide--again, lethality as microcolonies. DNA metabolism checkpoints. First identified by mutants that failed to respond properly to radiation (many are called "rad" mutants), or treatment with the drug hydroxyurea, which blocks at an early stage of S phase by inhibiting ribonucleotide reductase o While at first it was thought there was a single response pathway, subsequently became apparent that HU and radiation and other forms of damage challenge the cell in subtly different ways. Referred to as replication and damage checkpoints, respectively o These pathways overlap and use conserved kinases. General scheme can be defined as shown at right. However, there are differences in the details in different species. o In mammals, two kinases at top of pathway ATM and ATR which respond to different signals. In cerevisiae, major kinase homologue is MEC1. Similarly, in pombe, Rad3 is the major player. The yeast kinases appear to respond to all types of signal. Downstream, the Chk2 kinase is called RAD53 in budding yeast, Cds1 in fission yeast. Fortunately, Chk1 has the same name in all species. o Different pathways activated in response to different challenges. The Chk2/RAD53/Cds1 pathway responds early; the Chk1 pathway is specific for damage later in the cell cycle. As well as arresting cell cycle progression, the Chk2/RAD53/Cds1 pathway actively promotes repair of the damage. In mammalian cells, p53 is an important player but there is no p53 equivalent in yeast. Apoptosis is also regulated by this pathway; again, there is no similar phenomenon in single celled yeast. Experiments in fission yeast help to elucidate the signal: o Rad3 activates the downstream kinases Chk1 and Cds1/RAD53/Chk2 in response to different signals. o Chk1 is the damage-response kinase. It is activated by DNA damage, irradiation, or S phase progression (during which cells generate chromosomal structures characteristic of DNA damage). It phosphorylates Cdc25 protein which is inactivated and also exported from the nucleus. Chk1 mutants are viable, but sensitive to irradiation and other forms of damage. o Cds1/RAD53/Chk2 is the replication-response kinase. While it may overlap with chk1, its primary function appears to be in regulating replication in response to perturbations. For example, in HU treated cells, Cds1/RAD53 is active, and prevents the cells from firing/extending their replication forks while HU is present. It is also required for cells to be able to re-start replication when HU is removed and to repair DNA damage. Cds1/RAD53 checkpoint mutants are viable, and sensitive to HU. They are not particularly radiation sensitive. o Cells that fail to initiate S phase never activate the checkpoint, because they never send the signal that replication has started. Thus, initiation mutants such as cdc18-null in S pombe try to divide without replicating, generating a cut phenotype (see phenotypes, above). In contrast, cells that initiate, but cannot complete S phase, do activate the checkpoint. Classic cdc-style arrest of replication mutants is therefore checkpoint-dependent. o What happens to cells lacking checkpoint? No effect, as long as nothing goes wrong. BUT cannot arrest mitosis when damaged. Cannot prevent mitosis when S phase does not occur o Some mutants are checkpoint-defetive because they do not initiate the signal, such as cdc18-null. some are bona fide checkpoint mutants, such as rad3 or chk1 mutants. some are defective in the receiver. For example, cdc2-3w, version of cdc2 that is independent of cdc25 activation, is naturally checkpoint deficient. Ditto cdc2-Y15F. o A conserved group of proteins called the "checkpoint rads" may be involved in sensing the damage and activating Rad3. These have similarity to DNA replication proteins. During DNA replication, the PCNA protein is clamped around the DNA by its loading factor, RFC1-5. Intriguingly, the checkpoint Rad17 prtoein is structurally related to the RFC1 replication protein), and Rad1, Rad9, and Hus1 form a complex that looks like PCNA. Current models suggests that Rad17 and the RFC2-5 subunits form a damage-specific clamp loader protein that loads on the Rad1/9/Hus1 complex which in some way signals damage to Rad3. However, not all forms of damage require the checkpoint rads for the activation of Rad3. in budding yeast, things are different. o damage checkpoint still regulates mitosis, but by modulating activity of APC and its targets, instead of Cdc2. That is, regulates events in mitosis rather than events pre-mitosis. This may reflect very short G2 in budding yeast (S and M phase normally tightly coupled). Thus, PDS1 is a crucial molecule in the replication and damage checkpoint o responses in budding yeast. Also, the MEC1 and RAD53 proteins are essential for viability, indicating an essential role for these proteins. However, there are alleles that are checkpoint defective but viable, indicating the essential and checkpoint functions are genetically separable. In humans, these proteins are also present. Significantly there are additional transducers, including p53 and BRCA tumor suppressor genes, which are substrates of the ATM/ATR (=Rad3) family of kinases. o p53 highly unstable, targeted for destruction by binding MDM2. This binding prevented when p53 phosphorylated by checkpoint kinase cascade. o p53 induces expression of range of genes including CKIs that block CDK activity o Inactivation of o CDKs blocks engine of cell cycle, including preventing Rb from activating S phase genes. Visit this site for more about p53. spindle checkpoint: separate mechanism monitors whether cells assemble their chromosomes and spindles correctly. This works by affecting APC activity, in all systems. Don't want to divide in absence of aligned chromosomes, or might lose one. In yeasts this pathway is not essential for life, but in other organisms, it is required. This may indicate that more complex genomes are more prone to problems. o pathway bifurcates. One arm: Mad2 protein binds to unattached kinetochores, and prevents APC activation by binding Cdc20 (APC activating complex). Other arm: Bub2 inhibits activation of TEM1 and blocks activity o o of the Mitotic Exit network (MEN). Once all kinetochores attached and spindle is ready, MAD/BUB release blocks of APC/MEN and mitosis can proceed. Unattached kinetochores are phosphorylated, which may lead to Mad2 binding. Mad1 binds Mad2 and is a substrate for the MPS1 kinase. Mad2 binds CDC20, which prevents APC activation. BUB2 is also a substrate for MPS1. As described above, BUB2 prevents the GTPase TEM1 from being activated and thus blocks activation of the Mitotic Exit Network. This keeps the mitotic cyclins from being degraded, and thus keeps cerevisiae cells in mitosis. In fission yeast, the BUB2 o 12). Meiosis equivalent prevents cells from undergoing cytokinesis. This occurs during G1. Thus, the checkpoint response, like the APC pathways, vary in different organisms although they use conserved proteins. MAD and BUB proteins involved in all these checkpoints in yeast are also involved in human cells. As is the case for DNA metabolism checkpoints, in some cases there are several related proteins where yeast has one. Ability to manipulate spindle dynamics in larger cells allows testing of theories from genetical yeast expts. Meiosis is a specialized cell cycle that functions to reduce a diploid to a haploid. This is accomplished by undergoing one round of S phase followed by two divisions, reducing a diploid to 4 haploids in g1 phase. For basic description of meiosis including detailed diagrams, go to this page on this site. Many of the "usual" cell cycle proteins are involved in meiosis as well, including the CDKs, cyclins, APC, and cohesins. There appear to be meiosis-specific versions of at least some of these proteins functioning in addition to their mitotic counterparts. A central component to meiosis is the recombination between homologous chromosomes, which is required for proper chromosomal segregation. Recombination appears to require DNA replication and recombination proteins may influence the rate of meiotic progression. Additionally, DNA replication in meiosis may involve different proteins from those involved in vegetative cell, at least those involved in origin control. One particular area of interest recently in cohesion. There is a meiosis-specific cohesin, Rec8, that appears to be important for orienting the kinetchores properly and allowing the reductional and equational divisions to occur properly. o In the meiosis I division, the homologous chromosomes separate, but the sister chromatids remain attached. This is the reductional division, because it essentially reduces the number of chromosomes in the daughter nuclei from two to one. Recombination intermediates hold the chromosomes in synapsis, presumably limiting the access of the spindle to one kinetechore per homologue. During MI, the cohesion dissolves in the chromosome arms, but remains around the centromeres. Thus, the homologues remain attached, but the recombined arms can separate from each other. o During the MII division, the homologues separate, much as they do in mitosis in vegetative cells. In MII, the remaining cohesin around the centromeres is degraded so that the chromosomes separate properly. Other sites Cell cycle slide show Visit Mitosis World The Cell Cycle CellsAlive.com has excellent schematics of animal cell mitosis and the cell cycle Quiz yourself on mitosis and meiosis Cell Cycle Chapter of MOLECULAR BIOLOGY OF THE CELL (on line at NCBI) Sample topics including CDK and cyclins, from a textbook called The Cell Cycle: Principles of Control. YOu can get started here Milestones in the cell division from Nature More cell cycle notes/study tips Cell cycle and cell division links What the heck are all these gene names anyway? More than many fields, the cell cycle is particularly complicated because of the plethora of different gene names in different systems. One option in lecture is to use just one generic name--but then you can't read any papers, because everyone in the literature uses different gene names. Here is a table that should help negotiate the different species and different nomenclatures in this lecture. Factor what is it S. S. cerevisiae pombe metazoans CDK cyclin dependent kinase CDC28 Cdc2 Multiple CDKs: CDK16 G1 cyclin regulatory subunit of CDK for cell cycle entry CLN1,2 and 3 ? Cdk4-cyclinD S phase cyclin regulatory subunit of CDK for S phase entry CLB5, 6 Cig2 Cdk2-cyclinE late S phase cyclin regulatory subunit of CDK for S phase progression CLB3, 4 ? Cdk2-cyclinA M phase cyclin regulatory subunit of CDK for mitosis CLB1, 2 Cdc2<CDK1)Cdc13 CYCLINB< TD> APC Multi-component ubiquitin ligase required for degradation of substrates in mitosis and G1 Many genes Many genes Many genes APC specificity factors target the APC towards different substrates CDC20 HCT1 Slp1 Srw1 Cdc20, fizzy Hct1, Fzr securin An APC target, inhibits sister chromatid separation PDS1 Cut2 securin separase The securin target, a protease that degrades cohesin ESP1 cut1 separase Cohesin A complex of proteins that holds sister chromatids together SCC1, aka MCD1 SCC3 SMC1 SMC3 Rad21 aka Rad21 SCC1 psc3 SCC3 psm1 SMC1 Psm3 SMC3 SCF Multi-component ubiquitin ligase required for degradation of phosphorylated substrates in G1 SKP1 Cdc53 Cdc4 S is SKP1 SKP1 C is cullin ? F is F box Pop1, 2 protein CKIs CDK inhibitors--generally small SIC1 molecules, not conserved in primary Rum1 p16 p19 sequence p21 p27 ATM/ATR Master kinase regulators of checkpoint pathways checkpoint sensor RAD24 Complex of proteins consisting of a MEC3 clamp loader and a clamp that binds RAD17 DNA and monitors damage DDC1 Effector kinases Downstream of sensor kinase, respond to different challenges CHK1 (damage) CHK1 CHK1 RAD53 Cds1 CHK2 (HU) MEN Mitotic exit network, regulates progression out of M phase in S. cerevisiae. Similar proteins in S. pombe regulate septation (called SIN, for septation initiation network). Contains a GTPase, 2component GTP exchange factor (GEF), and a GAP, upstream of a phosphatase (PPase) Regulated (in S. cerevisiae) by nucleolar localization via protein Net1 TEM1 (GTPase) BUB2 BYR4 LTE1 CDC14 (PPase) Net1 Tumor suppressors Negative regulators of the cell cycle, NONE which are not found in fungi Regulated transcription; the ones Transcription here are active for synthesis of S factors phase genes MEC1 TEL1 SWI6 SWI4 MBP1 Rad3 Tel1 ATR ATM Rad17 Hus1 Rad1 Rad9 Rad17 Hus1 Rad1 Rad9 Spg1 Cdc16 Byr4 ? ? Clp1 ? NONE p53 Rb Cdc10 E2F Res1 preRC Orp1-6 Pre-replication complex, which ORC1-6 ORC1-6 Cdc18 marks a replication origin as ready to CDC6 CDC6 Mcm2fire MCM2-7 MCM2-7 7 Cdc7 Origin-activating kinase, which may play other roles in maintaining CDC7 genome integrity. Requires a subunit DBF4 (DBF4) which does not look like, but acts like, a cyclin Hsk1 Dfp1 CDC7 DBF4/ASK1 The Cell Cycle | Evidence for Regulation | Genetic Analysis | Cdc2 Regulation | Cyclins | Inhibitors | Destruction | Mitotic exit | Replication | S to M phase | Checkpoints | Meiosis Created 4/00 Last updated 041502 text and original drawings © S. L Forsburg Made on a Macintosh.