Plasmodium
advertisement

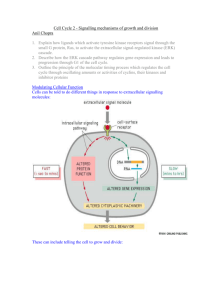
Plasmodium falciparum protein kinases: from database mining to drug discovery Inserm team (U609) Wellcome Centre for Molecular Parasitology University of Glasgow U609 Outline 1. Protein kinases 2. Atypical features of malarial protein kinases and signalling pathways: - CDKs - cyclins - MAPK pathways - Kinome 3. Malarial protein kinases as drug targets - target validation / functional studies - screening against recombinant targets - perspectives outline Protein phosphorylation: - involved in most cellular regulatory processes - 30% of proteins are phosphoproteins - major mechanism of signal transduction Protein kinases: 125 in yeast (ser/thr) 520 in human (ser/thr + tyr) (approx 2% of genes) Dysregulation associated with a number of pathologies (cancer, neurodegenerative diseases…) Kinase inhibitors on the market (Gleevec) or in clinical trials targets in parasitic diseases??? Introduction Structure of Ser/Thr and Tyr protein kinases GxGxxG K E I II III HRDxxxxN IV V VIA VIB DFG APE D VII VIII IX R X XI Kinase assay N-ter lobe Protein kinase ATP P C-ter lobe ATPPP Human CDK2 ADPP Introduction Introduction “…from yeast to man” Plasmodium Baldauf, Science 300, 1703 (June 2003) Introduction Outline 1. Protein kinases 2. Atypical features of malarial protein kinases and signalling pathways: - CDKs - cyclins - MAPK pathways - Kinome 3. Malarial protein kinases as drug targets - target validation / functional studies - screening against recombinant targets - perspectives outline Eukaryotic cell cycle pheromone Heterotrimeric G-proteins receptor CDK7 Cyclin H Ste20 Ste5 KSS1 Ste11 Ste7 Fus3 CDK4 Cyclin D CDK1 Cyclin B (MAPKKK) (MAPKK) (MAPK) CDK6 Cyclin D CDK3 Cyclin ? Cyclin A CDK1 CDK2 Cyclin E CDK2 Cyclin A CDKs CDKs of P. falciparum PSTAIRE T161 Cdk1 PSTTIRE T161 PfPK5 (Cdk1) AKTYIRE T161 Pfcrk-3 (Cdk1) KCILRE T161 PfPK6 (Cdk1/MAPK) Cyclin E Cyclin A Cyclin A CDK2 CDK1 S CDK2 M Pfcrk-4 (Cdk1/MAPK) CDK1 CDK3 CDK6 Cyclin D T161 Cyclin B G1 Cyclin ? EEFAVNE G2 CDK4 Cyclin D CDK7 Cyclin H Cell cycle control elements AMTSLRE T161 Pfcrk-1 (p58) NFVLLRE T161 Pfmrk (CDK7) ...and four potential cyclins CDKs Activation of PfPK5 (CDK1) by human cyclin H Pfmrk + PfPK5 + hcycH hcycH Pfmrk hcycH Pfmrk histone H1 Relaxed cyclin specifity for PfPK5? CAK-independent activation PfPK5 + Pfmrk + hcycH PfPK5 Histone H1 Human cyclin H Pfcyc-1, -3 p25 RINGO (A. Nebreda) Human cyclin A (J. Endicott) Autophosphorylation of PfPK5 Le Roch et al. (2000), JBC 275, 8952; Merckx et al., JBC (2003) CDKs PfPK5: CDK Pfmap-1: MAPK PfPK6: CDK-MAPK? sw|Q07785|CC2H_PLAFK CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (E... 592 e-169 sw|P43063|CC28_CANAL CELL DIVISION CONTROL PROTEIN 28 (EC 2.7.1... 194 3e-49 sw|Q11179|YPC2_CAEEL PUTATIVE SERINE/THREONINE-PROTEIN KINASE C... 328 1e-89sw|P23437|CDK2_XENLA CELL DIVISION PROTEIN KINASE 2 (EC 2.7.1.-... 192 8e-49 sw|P34117|CC2H_DICDI CDC2-LIKE SERINE/THREONINE-PROTEIN KINASE ... 411 e-115 sw|P28869|ERK1_CANAL EXTRACELLULAR SIGNAL-REGULATED KINASE 1 (E... 254 2e-67 sw|P29618|CC21_ORYSA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 1 ... 382 e-106 sw|Q39026|MPK6_ARATH MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 6... 190 2e-48 sw|P27638|SPK1_SCHPO MITOGEN-ACTIVATED PROTEIN KINASE SPK1 (EC ... 254 2e-67 sw|Q63699|CDK2_RAT CELL DIVISION PROTEIN KINASE 2 (EC 2.7.1.-). 190 2e-48 sw|Q38772|CC2A_ANTMA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG A ... 379 e-105 EXTRACELLULAR SIGNAL-REGULATED KINASE 1 (E... 253 5e-67sw|P23573|CC2C_DROME CELL DIVISION CONTROL PROTEIN 2 COGNATE (E... 190 2e-48 sw|P23111|CC2_MAIZE CELL DIVISION CONTROL PROTEIN 2 HOMOLOGsw|P42525|ERK1_DICDI (EC... 379 e-105 sw|Q39024|MPK4_ARATH MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 4... 251 2e-66 sw|P24100|CC2A_ARATH CELL DIVISION CONTROL PROTEIN 2 HOMOLOG A ... 374 e-103 sw|P24941|CDK2_HUMAN CELL DIVISION PROTEIN KINASE 2 (EC 2.7.1.-... 190 3e-48 sw|Q39025|MPK5_ARATH MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 5... 248 1e-65 sw|P52389|CC2_VIGUN CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC... 372 e-103 sw|O55076|CDK2_CRIGR CELL DIVISION PROTEIN KINASE 2 (EC 2.7.1.-). 190 4e-48 sw|Q07176|MMK1_MEDSA MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG M... 247 sw|P48963|CDK2_MESAU 2e-65 sw|P29619|CC22_ORYSA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 2 ... 370 e-102 CELL DIVISION PROTEIN KINASE 2 (EC 2.7.1.-). 188 1e-47 MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG M... 247 sw|P43450|CDK2_CARAU 3e-65 sw|P43450|CDK2_CARAU CELL DIVISION PROTEIN KINASE 2 (EC 2.7.1.-).sw|Q40353|MMK2_MEDSA 369 e-102 CELL DIVISION PROTEIN KINASE 2 (EC 2.7.1.-). 188 1e-47 MITOGEN-ACTIVATED PROTEIN KINASE 3 (EC 2.7... 247 3e-65 sw|Q39023|MPK3_ARATH MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 3... 187 3e-47 sw|Q63699|CDK2_RAT CELL DIVISION PROTEIN KINASE 2 (EC 2.7.1.-). sw|P27361|MK03_HUMAN 367 e-101 MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG N... 246 5e-65 sw|O55076|CDK2_CRIGR CELL DIVISION PROTEIN KINASE 2 (EC 2.7.1.-). sw|Q40531|NTF6_TOBAC 367 e-101 sw|Q07176|MMK1_MEDSA MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG M... 187 3e-47 sw|P47812|MK14_XENLA MITOGEN-ACTIVATED PROTEIN KINASE 2 (EC 2.7... 246 5e-65 sw|Q40532|NTF4_TOBAC MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG N... 186 6e-47 sw|P24941|CDK2_HUMAN CELL DIVISION PROTEIN KINASE 2 (EC 2.7.1.-... 366 e-101 MITOGEN-ACTIVATED PROTEIN KINASE 3 (EC 2.7.1... 246 6e-65 sw|P54666|CC23_TRYBB CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 3 ... 185 1e-46 sw|P48963|CDK2_MESAU CELL DIVISION PROTEIN KINASE 2 (EC 2.7.1.-).sw|P21708|MK03_RAT 366 e-101 sw|Q06060|MAPK_PEA MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG D5 ... 245 8e-65 sw|Q05006|CC22_MEDSA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 2 ... 365 e-101 sw|Q06060|MAPK_PEA MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG D5 ... 185 1e-46 MITOGEN-ACTIVATED PROTEIN KINASE (EC 2.7.1... 245 1e-64 sw|P24100|CC2A_ARATH CELL DIVISION CONTROL PROTEIN 2 HOMOLOG A ... 184 2e-46 sw|P23437|CDK2_XENLA CELL DIVISION PROTEIN KINASE 2 (EC 2.7.1.-...sw|P26696|MK01_XENLA 360 2e-99 sw|Q39026|MPK6_ARATH MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 6... 245 1e-64 sw|Q00526|CDK3_HUMAN CELL DIVISION PROTEIN KINASE 3 (EC 2.7.1.-). 356 2e-98 sw|P23111|CC2_MAIZE CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC... 184 2e-46 sw|Q39021|MPK1_ARATH MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 1... 243 3e-64 sw|P24923|CC21_MEDSA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 1 ... 356 4e-98 sw|P29618|CC21_ORYSA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 1 ... 183 4e-46 sw|Q40532|NTF4_TOBAC MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG N... 243 4e-64 sw|Q00535|CDK5_HUMAN CELL DIVISION PROTEIN KINASE 5 (EC 2.7.1.-... 355 5e-98 sw|P39951|CC2_RAT CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC 2... 182 7e-46 MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 7... 243 4e-64 sw|Q02399|CDK5_BOVIN CELL DIVISION PROTEIN KINASE 5 (EC 2.7.1.-...sw|Q39027|MPK7_ARATH 355 7e-98 sw|Q38772|CC2A_ANTMA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG A ... 182 7e-46 MITOGEN-ACTIVATED PROTEIN KINASE SUR-1 (EC... 243 5e-64 sw|P06493|CC2_HUMAN CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC... 182 9e-46 sw|P51166|CDK5_XENLA CELL DIVISION PROTEIN KINASE 5 (EC 2.7.1.-...sw|P39745|SUR1_CAEEL 354 1e-97 MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG N... 242 7e-64 sw|P49615|CDK5_MOUSE CELL DIVISION PROTEIN KINASE 5 (EC 2.7.1.-...sw|Q40517|NTF3_TOBAC 354 1e-97 sw|P48734|CC2_BOVIN CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC... 181 1e-45 sw|P27703|MK01_MOUSE MITOGEN-ACTIVATED PROTEIN KINASE 1 (EC 2.7... 242 7e-64 sw|P34117|CC2H_DICDI CDC2-LIKE SERINE/THREONINE-PROTEIN KINASE ... 181 2e-45 sw|P43063|CC28_CANAL CELL DIVISION CONTROL PROTEIN 28 (EC 2.7.1... 353 3e-97 sw|P28482|MK01_HUMAN MITOGEN-ACTIVATED PROTEIN KINASE 1 (EC 2.7... 242 7e-64 sw|P52389|CC2_VIGUN CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC... 180 3e-45 sw|Q38773|CC2B_ANTMA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG B ... 353 3e-97 MITOGEN-ACTIVATED PROTEIN KINASE 1 (EC 2.7... 242 7e-64 sw|P11440|CC2_MOUSE CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC... 180 3e-45 sw|Q03114|CDK5_RAT CELL DIVISION PROTEIN KINASE 5 (EC 2.7.1.-) ... sw|P46196|MK01_BOVIN 352 6e-97 sw|Q39022|MPK2_ARATH MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 2... 242 9e-64 sw|P06493|CC2_HUMAN CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC... 351 1e-96 sw|P34556|CC2_CAEEL CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC... 180 3e-45 sw|Q40884|MAPK_PETHY MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 1... 242 1e-63 sw|P39951|CC2_RAT CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC 2... 351 1e-96 sw|Q04770|CDK2_ENTHI CELL DIVISION PROTEIN KINASE 2 HOMOLOG (EC... 179 6e-45 sw|Q63844|MK03_MOUSE MITOGEN-ACTIVATED PROTEIN KINASE 3 (EC 2.7... 240 3e-63 sw|P11440|CC2_MOUSE CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC... 350 2e-96 sw|Q05006|CC22_MEDSA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 2 ... 179 7e-45 MITOGEN-ACTIVATED PROTEIN KINASE SLT2/MPK1... 240 5e-63sw|P29619|CC22_ORYSA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 2 ... 178 1e-44 sw|P48734|CC2_BOVIN CELL DIVISION CONTROL PROTEIN 2 HOMOLOGsw|Q00772|SLT2_YEAST (EC... 350 2e-96 MITOGEN-ACTIVATED PROTEIN KINASE 14 (EC 2.7.... 239 8e-63 sw|Q40517|NTF3_TOBAC MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG N... 178 2e-44 sw|P51958|CC2_CARAU CELL DIVISION CONTROL PROTEIN 2 HOMOLOGsw|P70618|MK14_RAT (EC... 348 7e-96 MITOGEN-ACTIVATED PROTEIN KINASE 14 (EC 2.... 239 8e-63 sw|Q39025|MPK5_ARATH MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 5... 178 2e-44 sw|P97377|CDK2_MOUSE CELL DIVISION PROTEIN KINASE 2 (EC 2.7.1.-).sw|P47811|MK14_MOUSE 344 2e-94 MITOGEN-ACTIVATED PROTEIN KINASE 14 (EC 2.... 239 8e-63 sw|Q40884|MAPK_PETHY MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 1... 178 2e-44 sw|P13863|CC2_CHICK CELL DIVISION CONTROL PROTEIN 2 HOMOLOGsw|Q16539|MK14_HUMAN (EC... 343 3e-94 sw|Q90336|MK14_CYPCA MITOGEN-ACTIVATED PROTEIN KINASE 14 (EC 2.... 239 8e-63 sw|P13863|CC2_CHICK CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC... 178 2e-44 sw|P35567|CC21_XENLA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 1 ... 341 1e-93 sw|O02812|MK14_CANFA MITOGEN-ACTIVATED PROTEIN KINASE 14 (EC 2.... 238 1e-62 sw|Q07785|CC2H_PLAFK CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (E... 177 2e-44 sw|P24033|CC22_XENLA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 2 ... 340 1e-93 MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 3... 238 2e-62 sw|P48609|CDK5_DROME CELL DIVISION PROTEIN KINASE 5 HOMOLOGsw|Q39023|MPK3_ARATH (EC... 340 3e-93 sw|P24923|CC21_MEDSA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 1 ... 177 3e-44 sw|Q13164|MK07_HUMAN MITOGEN-ACTIVATED PROTEIN KINASE 7 (EC 2.7... 238 2e-62 sw|P24033|CC22_XENLA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 2 ... 176 5e-44 sw|P00546|CC28_YEAST CELL DIVISION CONTROL PROTEIN 28 (EC 2.7.1... 340 3e-93 MITOGEN-ACTIVATED PROTEIN KINASE 11 (EC 2.... 236 5e-62 sw|Q39027|MPK7_ARATH MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 7... 176 6e-44 sw|Q06309|CRK1_LEIME CELL DIVISION PROTEIN KINASE 2 HOMOLOG sw|O08666|MK11_MOUSE CRK... 339 3e-93 sw|P40417|ERKA_DROME MITOGEN-ACTIVATED PROTEIN KINASE ERK-A (EC... 232 8e-61 sw|P23572|CC2_DROME CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC... 339 4e-93 sw|P23572|CC2_DROME CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC... 176 6e-44 sw|Q92398|SPM1_SCHPO MITOGEN-ACTIVATED PROTEIN KINASE SPM1 (EC ... 231 2e-60sw|P35567|CC21_XENLA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 1 ... 176 6e-44 sw|P54664|CC21_TRYCO CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 1 ... 338 1e-92 MITOGEN-ACTIVATED PROTEIN KINASE MKC1 (EC ... 229 9e-60 sw|P34112|CC2_DICDI CELL DIVISION CONTROL PROTEIN 2 HOMOLOG sw|P43068|MKC1_CANAL (EC... 335 5e-92 sw|Q39021|MPK1_ARATH MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 1... 175 8e-44 MITOGEN-ACTIVATED PROTEIN KINASE KSS1 (EC ... 229 9e-60 sw|P51958|CC2_CARAU CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC... 175 8e-44 sw|P04551|CC2_SCHPO CELL DIVISION CONTROL PROTEIN 2 (EC 2.7.1.-...sw|P14681|KSS1_YEAST 335 9e-92 MITOGEN-ACTIVATED PROTEIN KINASE 12 (EC 2.7.... 228 1e-59 sw|P00546|CC28_YEAST CELL DIVISION CONTROL PROTEIN 28 (EC 2.7.1... 175 1e-43 sw|P54119|CC2_AJECA CELL DIVISION CONTROL PROTEIN 2 (EC 2.7.1.-...sw|Q63538|MK12_RAT 333 3e-91 MITOGEN-ACTIVATED PROTEIN KINASE FUS3 (EC ... 228 1e-59 sw|P04551|CC2_SCHPO CELL DIVISION CONTROL PROTEIN 2 (EC 2.7.1.-... 174 2e-43 sw|Q00646|CC2_EMENI CELL DIVISION CONTROL PROTEIN 2 (EC 2.7.1.-...sw|P16892|FUS3_YEAST 332 6e-91 sw|Q09892|STY1_SCHPO MITOGEN-ACTIVATED PROTEIN KINASE STY1 (EC ... 228 2e-59 sw|Q39022|MPK2_ARATH MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 2... 174 2e-43 sw|P38973|CC21_TRYBB CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 1 ... 332 6e-91 sw|Q15759|MK11_HUMAN MITOGEN-ACTIVATED PROTEIN KINASE 11 (EC 2.... 228 2e-59 sw|P34112|CC2_DICDI CELL DIVISION CONTROL PROTEIN 2 HOMOLOG (EC... 174 2e-43 sw|P17157|PH85_YEAST NEGATIVE REGULATOR OF THE PHO SYSTEM (EC 2... 328 6e-90 MITOGEN-ACTIVATED PROTEIN KINASE 12 (EC 2.... 227 3e-59 sw|Q00526|CDK3_HUMAN CELL DIVISION PROTEIN KINASE 3 (EC 2.7.1.-). 173 3e-43 sw|O35831|KPT2_RAT SERINE/THREONINE-PROTEIN KINASE PCTAIRE-2sw|O08911|MK12_MOUSE (E... 324 2e-88 sw|P53778|MK12_HUMAN MITOGEN-ACTIVATED PROTEIN KINASE 12 (EC 2.... 225 1e-58 sw|Q40353|MMK2_MEDSA MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG M... 173 4e-43 sw|P23573|CC2C_DROME CELL DIVISION CONTROL PROTEIN 2 COGNATE (E... 324 2e-88 sw|O15264|MK13_HUMAN MITOGEN-ACTIVATED PROTEIN KINASE 13 (EC 2.... 222 1e-57 sw|Q00532|KKIA_HUMAN SERINE/THREONINE-PROTEIN KINASE KKIALRE (E... 173 6e-43 sw|Q00537|KPT2_HUMAN SERINE/THREONINE-PROTEIN KINASE PCTAIRE-2 ... 323 4e-88 MITOGEN-ACTIVATED PROTEIN KINASE HOG1 (EC ... 220 5e-57 sw|Q92241|PH85_KLULA NEGATIVE REGULATOR OF THE PHO SYSTEM sw|Q92207|HOG1_CANAL (EC 2... 320 2e-87 sw|P32485|HOG1_YEAST MITOGEN-ACTIVATED PROTEIN KINASE HOG1 (EC ... 172 7e-43 sw|P32485|HOG1_YEAST MITOGEN-ACTIVATED PROTEIN KINASE HOG1 (EC ... 217 4e-56sw|Q15131|CDKA_HUMAN CELL DIVISION PROTEIN KINASE 10 (EC 2.7.1.... 172 7e-43 sw|Q04899|KPT3_MOUSE SERINE/THREONINE-PROTEIN KINASE PCTAIRE-3 ... 315 6e-86 PROBABLE SERINE/THREONINE-PROTEIN KINASE Y... 213 4e-55sw|Q39024|MPK4_ARATH MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG 4... 171 1e-42 sw|O35832|KPT3_RAT SERINE/THREONINE-PROTEIN KINASE PCTAIRE-3sw|P36005|KKQ1_YEAST (E... 314 2e-85 sw|P45983|MK08_HUMAN MITOGEN-ACTIVATED PROTEIN KINASE 8 (EC 2.7... 205 2e-52 sw|Q04899|KPT3_MOUSE SERINE/THREONINE-PROTEIN KINASE PCTAIRE-3 ... 171 1e-42 sw|Q00536|KPT1_HUMAN SERINE/THREONINE-PROTEIN KINASE PCTAIRE-1 ... 312 7e-85 sw|P49187|MK10_RAT MITOGEN-ACTIVATED PROTEIN KINASE 10 (EC 2.7.... 204 3e-52 sw|P24788|KP58_MOUSE GALACTOSYLTRANSFERASE ASSOCIATED PROTEIN K... 171 2e-42 sw|Q04735|KPT1_MOUSE SERINE/THREONINE-PROTEIN KINASE PCTAIRE-1 ... 310 2e-84 sw|P49185|MK08_RAT MITOGEN-ACTIVATED PROTEIN KINASE 8 (EC 2.7.1... 204 3e-52 sw|P39745|SUR1_CAEEL MITOGEN-ACTIVATED PROTEIN KINASE SUR-1 (EC... 171 2e-42 sw|Q07002|KPT3_HUMAN SERINE/THREONINE PROTEIN KINASE PCTAIRE-3 ... 307 1e-83 MITOGEN-ACTIVATED PROTEIN KINASE 10 (EC 2.... 201 1e-51 sw|Q92241|PH85_KLULA NEGATIVE REGULATOR OF THE PHO SYSTEM (EC 2... 171 2e-42 sw|P34556|CC2_CAEEL CELL DIVISION CONTROL PROTEIN 2 HOMOLOGsw|P53779|MK10_HUMAN (EC... 305 5e-83 sw|Q61831|MK10_MOUSE MITOGEN-ACTIVATED PROTEIN KINASE 10 (EC 2.... 200 3e-51 sw|Q40531|NTF6_TOBAC MITOGEN-ACTIVATED PROTEIN KINASE HOMOLOG N... 171 2e-42 sw|P54666|CC23_TRYBB CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 3 ... 300 2e-81 sw|P45984|MK09_HUMAN MITOGEN-ACTIVATED PROTEIN KINASE 9 (EC 2.7... 199 6e-51 sw|Q38773|CC2B_ANTMA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG B ... 171 2e-42 sw|P25859|CC2B_ARATH CELL DIVISION CONTROL PROTEIN 2 HOMOLOG B ... 293 2e-79 sw|P49186|MK09_RAT MITOGEN-ACTIVATED PROTEIN KINASE 9 (EC 2.7.1... 199 7e-51 sw|O35832|KPT3_RAT SERINE/THREONINE-PROTEIN KINASE PCTAIRE-3 (E... 170 3e-42 sw|Q38774|CC2C_ANTMA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG C ... 286 4e-77 sw|P41808|SMK1_YEAST SPORULATION-SPECIFIC MITOGEN-ACTIVATED PRO... 199 1e-50 sw|Q38775|CC2D_ANTMA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG D ... 284 1e-76 sw|P21127|KP58_HUMAN GALACTOSYLTRANSFERASE ASSOCIATED PROTEIN K... 170 3e-42 sw|P29619|CC22_ORYSA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 2 ... 194 3e-49 sw|Q00536|KPT1_HUMAN SERINE/THREONINE-PROTEIN KINASE PCTAIRE-1 ... 170 4e-42 sw|P54665|CC22_TRYBB CELL DIVISION CONTROL PROTEIN 2 HOMOLOG 2 ... 284 2e-76 sw|P40417|ERKA_DROME MITOGEN-ACTIVATED PROTEIN KINASE ERK-A (EC... 170 4e-42 CDKs HsCDK1 SpCdc2 PfPK5 PfCRK4 PfCRK4 PfPK6 PfPK6 MmErk2 ScFus3 PfMAP1 PfcPKA 702.2 500 200 100 50 20 10 5 2 1 CDKs MAPKs 0 CDKs MAP kinase pathway R TK ras MAP kinase kinase kinase MAP kinase kinase MAP kinase ? Cyclin E Cyclin A Cyclin A CDK2 CDK1 S G2 CDK2 M Cyclin B CDK1 G1 CDK3 PfPK6 Cyclin ? CDK6 Cyclin D Pfcrk-4 ? CDK4 Cyclin D CDK7 Cyclin H Cell cycle control elements CDKs Aim : To identify components of MAPK modules. pheromone G-proteins receptor Ste20 Two MAP kinase homologues Ste11 Ste7 KSS1 Fus3 Ste5 (MAPKKK) (MAPKK) (MAPK) F Y L K N C TXY F Y L K N C TXY F Y L K N C TSH Mammalian/yeast MAPK Pfmap-1 (IKEQ)5 (KKYVDGSN)12 Pfmap-2 MAPK pathway Activation of GST-Pfmap-2 by gametocyte extracts Activity of recombinant Pfmap-2 Pfmap-2 MBP % activity 600 400 200 100 Lane 1 2 3 4-5 6-7 Dorin et al. (1999), JBC 274, 29912. MAPK pathway Atypical MAPK pathway in Plasmodium Typical MAPK module P. falciparum MEKK SMANS TEY MEK1/2 MAPK (ERK1/2) ? TSH Pfmap-2 TDY Pfmap-1 MAPK pathway ? Can we use genomic databases to solve black boxes ? ? MAPK pathway …it works well for reconstruction of metabolic pathways... From Hagai Ginsburg’s web site on Plasmodium metabolic pathways (http://sites.huji.ac.il/malaria) « Metabolic databases have been used to predict the pathways of several organisms from their genomes. These concepts were at the base of the construction of this site ». MAPK pathway BLAST analysis using whole-length MEK1 as a query No hit in PlasmoDB BLAST analysis using 23 residues around the SMANS activation site 1 Hit! Query: 1 GVSGQL-IDSMANSFVGTRSYMSP 23 G+S + I+SMA+S VGT Y SP Subject: 591 GLSKNIGIESMAHSCVGTPYYWSP 662 30% 100% 42% Mm MEK1 IKLCDFGVS--GQLIDSMANSFVGTRSYMSPERLQGTHYSV--QSDIWSM : ::: : :.:::.: ::: : ::: : : :: : Pfnek-1 AKIGDFGLSK-NIGIESMAHSCVGTPYYWSPELLLHETKSYDDKSDMWAL : ::::::: : :::: : ::: : :: :: NIMA VKLGDFGLSKLMHSH-DF.STYVGTPFYMSPEIC--AAEKSTLRSDIWAV ( SMANS MEK1/2 ) SMAHS Pfnek-1 MAPK pathway Typical MAPK module P. falciparum MEKK SMANS MEK1/2 SMAHS Pfnek-1 ? TEY MAPK (ERK1/2) TSH Pfmap-2 TDY Pfmap-1 MAPK pathway Atypical MAPK pathway in Plasmodium Pfnek-1: - closely related to NIMA kinases; MAPKK–like activation site. SMANS MEK1/2 SMAHS Pfnek-1 (100%) DxxT NIMA/Nek (42%) (30%) Phosphorylation of Pfmap-2 by Pfnek-1 Pfnek-1: ERK2 Pfmap-2 + - + - + Autoradiogram Synergistic activities of Pfnek-1 and Pfmap-2 mERK2 Pfmap-2 Coomassie mERK2 map-2 nek-1 nek-1 + map-2 MBP Pfmap-2 Dorin et al. (2001), EJB 268:2600-2608 MAPK pathway Aim : To identify components of MAPK modules. pheromone G-proteins receptor No typical MAPKK homologues! Ste20 Ste11 Ste7 KSS1 Fus3 Ste5 - Experimental (PCR) approaches - Targeted database mining (BLASTP) (MAPKKK) (MAPKK) (MAPK) - Genome-wide “kinome” characterisation (with P. Ward, WCMP, and J. Packer, Abbott laboratories) PlasmoDB database mining using a variety of tools (HMM, BLAST, Gene Ontology prediction..) Alignment, manual edition Construction of a tree including members of defined families of eukaryotic protein kinases Kinome Features of the P. falciparum kinome: • Only 65 „typical“ S/T protein kinases (124 in yeast) • Many can be attributed to distinct families (35-60% identity) : - 2 MAPKs, 6 CDK-related kinases, PKA, PKG, PKB... - at least 5 CDPKs (found also in plants and Ciliates, but not in animal cells) • Many do not cluster with previously defined kinase groups • Several show features from distinct kinase families • No TyrK, no PKC, no STE Kinome pfb0150c map2k1 map3k3 map4k1 pak1 csnk1g1 Pf110377 (PfCK1) csnk1d Casein kinase 1 Ste7/11/20 pf140320 pf110220 mal6p1191 pf130258 tgfbr1 Pfb0520w (PfRaf) zak limk1 raf1 src insr egfr pfi1280c pfi1290w Mal6p1146 (PfPK4) pfa0380w pf140423 pfl0080c mal6p156 Pfl1370w (Pfnek-1) nek7 Pfe1290w nek1 Mal7p1100 (Pfnek-4) Mal7p191 (PfEST) pf110488 mal13p1196 Pfc0755c (Pfcrk-4) Mal6p1271 (Pfcrk-5) Pf130206 (PfPK6) Pfd0740w (Pfcrk-3) Mal13p1279 (PfPK5) cdk2 Pf100141 (Pfmrk) Pfd0865c (Pfcrk-1) Pf110147 (Pfmap-2) Pf140294 (Pfmap-1) mapk1 Pf110096 (PfCK2) Pf080044 (PfPK1) mal13p184 Pfc0525c (PfGSK3) gsk3b pf110156 Pf140431(Pflammer) clk1 pfc0105w pf140408 pf140392 mal7p173 Pfl1885c (PfPK2) pf110242 Pfb0815w (PfCDPK1) pf070072 Pf130211 Mal6p1108 (PfCDPK2) Pfc0420w (PfCDPK3) mapkapk2 camk2a camk1 dapk1 pf110239 pf140227 pfc0485w pfb0665w mal7p118 pf110060 pf110464 pfc0385c mal13p1278 pf110227 Pf140346 (PfPKG) rock1 Pfl2250c prkca akt1 prkaca Pfi1685w (PfPKA) Pf140516 (PfKIN) Pf140476 pf130085 Pfb0605w (PfPK7) Pfl2280w Tyrosine kinase-like Tyrosine kinase NimA CMGC Camk AGC Kinome pfi1415w A novel, Apicomplexa-specific family of PK-like proteins GxGxxG K E I II III W HRDxxxxN IV V FIKKxxxxxW GExxxxxxNxxMExxxxxxL VIA VIB LHxxGxxHLDxxxxNxL DFG APE D VII VIII IX PxPE YxxVxxxDxxM R X WP XI LxxxxxR PW CD[FL][GA]xxxxxPxY • 20 genes in P. falciparum • 1 gene in P. vivax and P. berghei • 1 gene in Toxoplasma gondii and Cryptosporidium • Not found so far in any non-apicomplexan organism • Closest relatives (by BLASTP !): MAPKs Kinome PfEMP1 (var) rifin FIKK Kinome the organisation of Plasmodium regulatory pathways cannot be predicted from sequence databases by analogy to those in higher Eukaryotes. Kinome Reconstructing metabolic pathways Model eukaryote metabolites Plasmodium Enzyme A Enzyme B Enzyme C Kinome Problems in reconstructing regulatory pathways: recognisable not recognisable Kinome Holton et al., Structure (2003) Kinome Outline 1. Protein kinases 2. Atypical features of malarial protein kinases and signalling pathways: - CDKs- cyclins - MAPK pathways - Kinome 3. Malarial protein kinases as drug targets - target validation / functional studies - screening against recombinant targets - perspectives outline Validation by gene knock-out Malaria parasites: - Extremely low transfection rate - No inducible expression system yet PyrR PyrS Pyr selection targeted gene Check for the presence of the desired genotype (PCR) PyrR Clone parasites Analyse phenotype Kinases as drug targets Pbcrk-1 cannot be knocked-out… MscI MscI 1734 Pbcrk-1 202 Pst I 1027 MscI hDHFR Homologous recombination MscI MscI MscI episome Rangajaran et al., submitted WT Kinases as drug targets …but the Pbcrk-1 locus is amenable to genetic modification. NdeI NdeI Pbcrk-1 802 XhoI NdeI 1734 hDHFR Homologous recombination NdeI hDHFR NdeI 1734 episome recombinant Rangajaran et al., submitted WT Kinases as drug targets Problem: detailed studies of the phenotype are impossible using KO approach. a possible solution: chemical genetics ? Validation by chemical genetics ATP Inhibitor ACTIVE Wild-type kinase Large gatekeeper: F, L... ATP Inhibitor INACTIVE Mutated kinase Small gatekeeper: G, A... Kinases as drug targets 338 V-Src C-Fyn C-Abl Cdk2 CDC28 CAMKII FUS3 332-EPIYIVIEYMSK--GSLLDFLKGEM333-EPIYIVTEYMNK--GSLLDFLKDGE309-PPFYIITEFMTY--GNLLDYLRECN83-NKLYLVFEFLHQ---DLKKFMDASA92-HKLYLVFEFLDL---DLKRYMEGIP84-GHHYLIFDLVTG--GELFEDIVARE87-NEVYIIQELMQT---DLHRVISTQMBishop et al., Nature 407:395-401 Pfmap-2 Pfmap-1 PfPK5 PfPK6 PfGSK-3 Etc... 171-DELYIVLEIADS---DLKKLFKTPI107-NDIYLIFDFMET---DLHEVIKADL73-KRLVLVFEHLDQ---DLKKLLDVCE109-SCLYLAFEYCDI---DLFNLIKKHN151-IFLNVVMEYIPQ---TVHKYMKYYS3. Parasite PKs as targets: validation Validation by chemical genetics Selective inhibition of mutagenised PfPK5-Pfcyc-1 by N-M-PP1 nM 1 N-M-PP1 Histone H1 100% 100% 50 50 His-PfPK5 WT His-PfPK5 F79G Next: allele replacement in the parasite Kinases as drug targets Chemical library screening Medium-throughput screening Preliminary inhibition assay (PfGSK3). 100% 90% 80% 70% 60% 50% 40% 30% 20% 10% 0% 0,01 A 0,1 1 10 Concentration (µM) Paullone Gwennpaullone 100 Kenpaullone Alsterpaullone next: - selectivity studies - cellular effects of the hits Kinases as drug targets Perspectives --Rational drug design Nu6102 binding to PfPK5 and Cdk2 • Mode of binding highly conserved from Cdk2 • Subtle differences in ribose binding site may afford greater potency for PfPK5 selective inhibitors •IC50: PfPK5, 215 nM CDK2, 5 nM Holton et al., Structure (2003) Kinases as drug targets In brief: 1. Parasite protein kinases display many divergences relative to their mammalian homologues. 2. Peculiarities of the malarial kinome: - absence of three-component MAPK modules - a novel family of kinase-related molecules - several „orphan“ kinases/groups (incl. „hybrid“ kinases) 3. Antimalarial drug discovery based on protein kinase inhibitors: - Screening on recombinant kinases is ongoing - Rational drug design in progress - Chemical genetics as a possible approach for functional studies Acknowledgements Jane Endicott, Univ. of Oxford Kavita Shah, Novartis, San Diego Laurent Meijer, CNRS, Roscoff/Rockefeller Debopam Chakrabarti, UCF, Orlando Robert Menard, Institut Pasteur Ali Sultan, Harvard School of Public Health Jeremy Packer, Abbott Laboratories WCMP-INSERM U609: Pauline Ward Dominique Dorin Karine Le Roch Anais Merckx Leila Equinet Marie-Paule Nivez Luc Reiniger Caroline Doerig