Data Analysis Pipeline from heterogeneous MS
advertisement

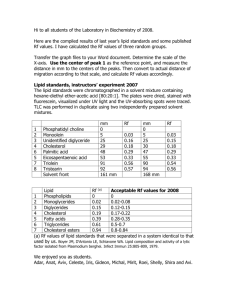
BIOM 209/CHEM 210/PHARM 209 Interrogating Gene, Protein and Lipid Databases: A Bioinformatics Perspective Dr. Eoin Fahy, University Of California San Diego ® Professor Edward A. Dennis Department of Chemistry and Biochemistry Department of Pharmacology, School of Medicine University of California, San Diego Copyright/attribution notice: You are free to copy, distribute, adapt and transmit this tutorial or individual slides (without alteration) for academic, non-profit and non-commercial purposes. Attribution: Edward A. Dennis (2010) “LIPID MAPS Lipid Metabolomics Tutorial” www.lipidmaps.org E.A. DENNIS 2016 © Definition of a lipid* Lipids may be broadly defined as hydrophobic or amphiphilic small molecules that originate entirely or in part from two distinct types of biochemical subunits or "building blocks": ketoacyl and isoprene groups. Using this approach, lipids may be divided into eight categories : fatty acyls, glycerolipids, ,glycerophospholipids, sphingolipids, saccharolipids and polyketides (derived from condensation of ketoacyl subunits); and sterol lipids and prenol lipids (derived from condensation of isoprene subunits). * Fahy,E. et al, Journal of Lipid Research, Vol. 46, 839-862, May 2005 Fundamental biosynthetic units of lipids Lipid classification: biosynthetic routes LIPID MAPS Classification System Categories and Examples Category Abbreviation Example Fatty acyls FA Dodecanoic acid Glycerolipids GL 1-hexadecanoyl-2-(9Z-octadecenoyl)-snglycerol Glycerophospholipids GP 1-hexadecanoyl-2-(9Z-octadecenoyl)-snglycero-3-phosphocholine Sphingolipids SP N-(tetradecanoyl)-sphing-4-enine Sterol lipids ST Cholest-5-en-3b-ol Prenol lipids PR 2E,6E-farnesol Saccharolipids SL UDP-3-O-(3R-hydroxy-tetradecanoyl)-aDN-acetylglucosamine Polyketides PK Aflatoxin B1 J. Lipid Res. Classification publications Journal of Lipid Research, Vol. 46, 839-862, May 2005 Journal of Lipid Research, 50th anniversary edition, May 2009 LIPID MAPS Lipid classification system Category Abbrev Fatty Acyls FA Glycerolipids GL Glycerophospholipids GP Sphingolipids Sterol Lipids Prenol Lipids Saccharolipids Polyketides SP ST PR SL PK Example Arachidonic acid 1-hexadecanoyl-sn-glycerol 1-hexadecanoyl-2-(9Z-octadecenoyl)sn-glycero-3-phosphocholine Sphingosine Cholesterol Retinol Kdo2-lipid A epothilone D Name: PGE2 LM_ID: LMFA03010003 LM_ID description: Database: LM (LIPID MAPS) Category: FA (Fatty Acyls) Main Class: 03 (Eicosanoids) Sub Class: 01 (Prostaglandins) Unique identifier within a sub class: 0003 LIPID MAPS: Recommendations for drawing structures Consistent structure representation across classes Fatty Acyls(FA) Glycerolipids (GL) Glycerophospholipids (GP) Sphingolipids (SP) Sterol Lipids (ST) Prenol Lipids (PR) Structural comparison of SM and PC Online lipid structure-drawing tools http://www.lipidmaps.org/tools/index.html Online drawing tools for various lipid categories (FA,GL,GP,SP,ST) Structures may be saved as Molfiles LIPID MAPS Lipidomics gateway http://www.lipidmaps.org #Lipids in LMSD by year 45000 40000 35000 30000 25000 20000 15000 10000 5000 0 2004 2005 2006 2007 2008 2009 2010 2011 2012 2013 2014 2015 Lipids per category in LMSD 10000 9000 Total: 40,360 8000 7000 6000 5000 4000 3000 2000 1000 0 FA GL GP SP ST PR SL PK Populating LIPID MAPS structure database Structures from core labs and partners New structures identified by LIPID MAPS experiments Computationally generated structures Public databases Websites, Publications LIPID MAPS structure database Search LMSD by browsing classification hierarchy Search LMSD by structure, text, mass, formula ,ontology Search LMSD with ontology terms e.g. find all lipids with 20 carbons, 3 double bonds, at least 3 hydroxyl groups and 1 epoxy group LMSD Detail view for a lipid structure Structure LM_ID Names, synonyms m/z calculation tool Lipid classification Database cross-references InChiKey identifier MS/MS spectrum Physicochemical properties Other structure formats Alternative lipid subclasses/functionality Take advantage of built-in ontology feature for all lipid structures in LMSD Use InChIKey to find structures differing only in stereochemistry, double-bond geometry or isotopic labeling Use InChIKey (full or partial) to perform a Google structure search European Bioinformatics Inst. LIPID MAPS PubChem Querying Lipidomics Gateway website as well as LIPID MAPS databases via “Quick search” Multi-purpose Small “footprint” High visibility (on home page) Search the Lipidomics Gateway html pages by keyword, or the databases by lipid class, common name, systematic name or synonym, mass, formula, InChIKey, LIPID MAPS ID, gene or protein term. Quick search query types LIPID MAPS LM_ID Lipid classification term Lipid common/systematic name or synonym Lipid molecular formula InChI Key Lipid standard (name or LMID) Gene/protein name/synonym Keywords on Lipidomics Gateway website pages (personnel, publications, news, updates, etc.) Example LMFA03010003 “Choline”, “prostaglandin”, “diterpene” “Linoleic”, “HETE”, ”, “PAF”, “PGE”, “5Z,8Z,14Z-eicosatrienoic”, “PC(16:0/18:1(9Z))” “MGDG “docosa”, “phytosphingosine” C12H24O2 XEYBRNLFEZDVAW-ARSRFYASSA-N sterol FABP “Atherosclerosis”, “Dennis”, “homeostasis” Lipid Proteome Database (LMPD) Species Genes Proteins Human (Homo sapiens) 1116 2273 Mouse (Mus musculus) 1082 1504 Rat (Rattus norvegicus) 1258 1315 Rhesus monkey (Macaca mulata) 891 1634 Yeast (Saccharomyces cerevisiae (s288c)) 720 720 E. coli (Escherichia coli(K12)) 245 245 C. elegans (Caenorhabditis elegans) 595 868 Drosophila (Drosophila melanogaster) 404 1064 1829 2447 638 647 Arabidopsis (Arabidopsis thaliana) Zebrafish (Danio rerio) LMPD:Data collection strategy Lipid-related keywords in gene names, metabolic pathways and ontology terms Manual curation Entrez Gene ID list NCBI Entrez Python program UniProt Gene, mRNA, protein data, PTM variants, motifs, homologs, crossreferences, related proteins, ontologies, annotations, etc. LMPD database LMPD organization: Gene-> mRNA-> (apo)protein -> mature protein Entrez Gene ID (DNA/genomic links) RefSeq mRNA ID’s (both coding and UTR variants) RefSeq protein ID’s and sequences (unique isoforms) Post–translationally modified variants (e.g. apo-, mature forms, leader sequences, etc.) LMPD query page LMPD overview page: listing of annotations and isoforms LMPD gene orthologs, alignments, links LMPD UniProt, domain/motif , related protein annotations LMPD gene ontology/pathway annotations LIPID MAPS REST interface LIPID MAPS REST interface Different input contexts: Compounds Genes Proteins Output formats: JSON, text, molfile, image JSON molfile LIPID MAPS lipidomic pathways Cholesterol Biosynthesis TLR4 signaling pathway Overview of Quantitative Lipid Analysis by Mass Spectrometry as performed by LIPID MAPS consortium on bone marrow derived macrophages (BMDM) www.lipidmaps.org LIPID MAPS funded by Glue Grant from :www.nigms.nih.gov Extract bone marrow cells Transfer to plates Repeat 3x (replicates) Perform timecourse experiment on plated cells Aliquot samples for shipping to core research labs Fatty acids Methanolic HCl/Isooctane extraction Eicosanoids Separate media from cells SPE extraction of media Glycerophospholipids Cardiolipins Glycerolipids Methanolic HCl/CHCl3 extraction Methanol/CHCl3 extraction EtOAc/isooctane extraction Cholesteryl esters EtOAc/isooctane extraction DFPI derivatization of DAGS Sterols Sphingolipids Saponification Methanol/CHCl3 + methanolic KOH extraction Methanol/CHCl3 extraction Prenols Methanol/CHCl3 extraction SPE extraction GC/MS analysis LC/MS analysis LC/MS analysis LC/MS analysis LC/MS analysis LC/MS analysis Deuterated standards (reverse phase) (normal phase) (normal phase) (normal phase) (normal phase) ESI-QTRAP viaMRM methods ESI-QTRAP 2-stage quantitation ESI-QSTAR-XL using MS/MS methods Deuterated standards Odd-chain standards Odd-chain standards ESI-QTRAP ESI-QTRAP [M+NH4]+ detection mode [M+NH4]+/neutral loss detection mode Deuterated standards Deuterated standards Combination of GC/MS, LC/MS (reverse phase) on ESI-QTRAP and APCIMS analysis Deuterated standards Combination of LC-C18, LC-Si and LC-NH2 separation ESI-QTRAP and API-3000 Triple Quad detection with MRM methods C12 analog standards LC/MS analysis (reverse phase) QSTAR-XL via MRM methods Nor-dolichol/CoQ6 standards BIOINFORMATICS Data consolidation, normalization, statistical analysis and databasing Presentation in tabular and graphical formats For details of extraction, purification and quantitation by MS, see: Lipidomics reveals a remarkable diversity of lipids in human plasma. Quehenberger O et al.,J Lipid Res 51, 3299-3305 (2010). A mouse macrophage lipidome. Dennis EA et al., J Biol Chem 285, 39976-39985 (2010) Methods Enzymol. (Brown AH, ed.) 2007; Vol. 432 (multiple chapters) LIPID MAPS Bioinformatics Corea, UCSD, 9500 Gilman Dr, La Jolla, CA 92093; Department of Bioengineeringb, UCSD, 9500 Gilman Dr, La Jolla, CA, 92093 Data presentation formats Graphical: Tabular: Heatmap: Lipids Integrated pathway/heatmap: Genes Dennis et al (2010) J. Biol. Chem, 51, 39976-85 E. Fahy 2010 © Online lipid structure-drawing tools http://www.lipidmaps.org/tools/index.html Online drawing tools for various lipid categories (FA,GL,GP,SP,ST) Structures viewable in Marvin, JMol and Chemdraw format. May be saved as Molfiles E. Fahy 2010 © Online generation of glycan structures in full chair conformation http://www.lipidmaps.org/tools/index.html Sugars Glc Gal GlcNAc GalNac Xyl Fuc Man NeuAc NeuGc KDN Anomeric Carbon a or b linkages may be specified E. Fahy 2010 © Mass spectrometry prediction tools Using virtual databases of structures based on commonly occurring core structures and chains Using known lipids in the LIPID MAPS structure database (LMSD) Creation of a virtual lipid database Choice of range of acyl/alkyl chains These are used to create “bulk” species e.g. PC(38:4), PE(O-36:0), Cer(d32:1), HexCer(d40:2), TG(54:2), DG(32:0), FA(20:3(OH)), CE(18:1) Conservative approach: stereochemistry, sn (glycerol) position, double bond/functional group regiochemistry, double bond geometry not defined. Links to: On-demand expansion of all possible chain combinations (within defined limits) Links to: Matches of bulk species to discrete structures in LMSD database (examples) Enumeration of “bulk” lipid species from selected lists of acyl/alkyl chains Suite of combinatorial expansion tools Glycerolipids Acyl CoA’s Phospholipids Acyl carnitines Cardiolipins Chol. esters Sphingolipids Wax esters Fatty acids Database of lipid “bulk” species, exact masses, formulae, annotations Virtual database of bulk lipids: number of entries per class Monoradylglycerols 84 Diradylglycerols 615 Triradylglycerols Fatty acids 13590 Acyl carnitines 78 1844 Chol. Esters 78 Digalactosyl DG's 553 Acyl CoA's 78 Monogalactosyl DG's 553 Wax esters 403 Sulfoquinovosyl DG's 553 Ceramides 258 PA 696 Ceramide phosphates 258 PC 696 PE-Ceramides 230 PE 696 PI-Ceramides 230 PG 696 Mannosyl-di-IP-ceramides 258 PI 696 Mannosyl-IP-ceramides 258 PIP 696 Hexosyl ceramides 258 PS 696 Lactosyl ceramides 258 Cardiolipins 375 Sphingomyelins 258 Sulfatides 258 Precursor ion search interface to virtual database Input: Either copy/paste a list of precursor ions or upload a peaklist file Input parameters: Mass tolerance, ion type, all chains or even chains, sort results Optionally restrict search to one or multiple lipid species Results page for precursor ion search Output: view in online format (below) or as tab-delimited text file Output features: Sub-table for each input ion. Links: On-demand expansion of all possible chain combinations (abbreviation) Links: Matches of bulk species to discrete structures in LMSD database (examples) Expansion of species level to display all possible chain combinations within defined chain and chain/double-bond ratio limits Links to examples of discrete structures in LMSD database with the identical bulk structure *This feature was implemented by computing the “bulk” abbreviation (where possible) for every structure in the LMSD database Educating the public about lipids Educating the public about lipids: LIPID MAPS tutorials http://www.lipidmaps.org LIPID MAPS ®