Tema3
advertisement

Tema 7: Homofermentative
Pathway
Chapter 14
Pages 383 - 402
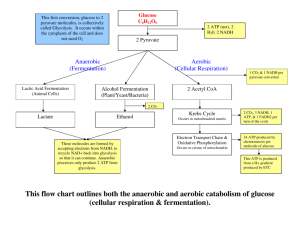
Formation of acetyl-CoA from pyruvate
Acetyl-CoA + CO2 + NADH2
Pyruvate
dehydrogenase
pyruvate
Pyruvate
Ferredoxin
oxidoreductase
Pyruvate
Formate lyase
Acetyl-CoA + formic acid
Acetyl-CoA + CO2 + H2
Anaerobically
Pyruvate
dehydrogenase
1) Catalyze an oxidative decarboxylation.
2) It is found in aerobically grown Bacteria, mitocondria, but
not in Archaea.
3) The product acetyl-CoA usually goes to the TCA cycle
instead of to acetyl-P
HSCoA
O
O
HOOC-C-CH3
NAD
CH3CO-SCoA + CO2
NADH2
TCA cycle
Pyruvate
Formate lyase
1) Catalyze an oxidative decarboxylation.
where the electrons remain in the carbonyl group.
2) The product acetyl-CoA usually goes to acetyl-P.
O
O
HOOC-C-CH3 + CoASH
CH3CO-SCoA + CH2O2
Pi
Phosphotransacetylase
ADP
CH3COOH + ATP
Mg+
O
CH3CO-P + HSCoA
Acetate kinase
1) Catalyze an oxidative decarboxylation where ferredoxin
is the Electron acceptor.
Pyruvate
Ferredoxin
oxidoreductase
2) It is found typically in clostridia and sulfate reducing bacteria
(SRB) and other anaerobes.
3) The product acetyl-CoA usually goes to acetyl-P.
O
O
HOOC-C-CH3 + CoASH
CH3CO-SCoA + CO2
Fd ox Fd red
Pi
hydrogenase
Phosphotransacetylase
2H+
ADP
CH3COOH + ATP
2H2
O
Mg+
CH3CO-P + HSCoA
Acetate kinase
How is acetyl-CoA made from Acetate?
It is typically made as follows
ADP
CH3COOH
OUT
O
Mg+
+ ATP
CH3CO-P + HSCoA
Acetate kinase
or
Phosphotransacetylase
acetylCoA
Pi
synthetase
O
IN
CH3CO-SCoA
Carbon and energy
Lactic Acid Bacteria
Characteristics: Gram positive, carbohydrate
users, proteolysis rare, nonmotile, non-spore
forming
Strict fermentors,
unable to synthesize cytochromes unless heme is
added.
catalase negative
oxidase negative
Nutritionally fastidious
All make lactic acid (lactate) as predominant
end product
Lactic Acid Bacteria
Types of fermentation
Homofermentative: glucose to 2 lactic acids,
85-95% of glucose carbon in lactate
Heterofermentative: glucose to 1 lactate, 1
ethanol, and 1 carbon dioxide, only 50% or
less of glucose carbon in lactate.
Types of products will define the pathway
used and ATP made.
Lactic Acid Bacteria
Types of organisms
Streptococcus: homofermentative
Leuconostoc: heterofermentative
Pediococcus homofermentative
Lactobacillus; heterofermentative or
homofermentative.
Lactic Acid Bacteria
Streptococcus species:
Enterococcus: gut dwellers
Lactococcus
natural fermentations
Lactic acid production: lowers pH,
preserves and precipitates proteins
Lactic acid bacteria
Homofermentative pathway
Uses Glycolytic pathway to make 2 pyruvates
from glucose
Overview:
Activation-use 2 ATP
Make ß-carbonyl
C-C bond cleavage
Oxidation/reduction
Substrate-level phosphorylation
Homofermentative Pathway in Streptococci
Glucose
ATP
ADP
Glucose-6-P
Fructose-6-P
ATP
ADP
Fructose-1,6 bis P
Dihydroxyacetone-P
2 Glyceraldehyde-3-P
2 NAD+
Dihydroxyacetone-P
2 NADH
converted to glyceraldehyde-3-P
2
1,3-bisphosphoglycerate
Pathway shows 2 G-3-P's after
this step.
2 ADP
2 ATP
2 3-phophoglycerate
Reoxidation of NADH
Lactate
dehydrogenase
2 pyruvate
2 NADH
2 2-phosphoglycerate
2 NAD+
2 lactate
2 H2O
2 phosphoenolpyruvate
2 ADP
Used 2 ATP
Made 4 ATP
Net ATP yield=2 ATP/glucose
2 ATP
2 pyruvate
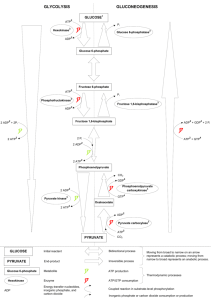
Glycolytic Pathway for Glucose Metabolism
CH2
HO
H
H
OH H
C
C
C
OH OH H
Hexokinase or PTS system
= OP
3
O
CH2
C
C
H
O
Glucose
OH
ATP
H
H
ADP
OH H
C
C
C
OH OH H
H
C
O
C
Glucose-6-P
OH
G6P isomerase
= OP
3
O
CH2
H
H
OH H
C
C
C
C
CH2OH
Fructose-6-P
OH OH H
O
ATP
Phosphofructokinase
= OP
3
O
CH2
Fructose-1,6-bis P
aldolase
= OP
3
O
CH2
H
H
ADP
OH H
C
C
C
C
OH OH H
O
H
H
C
C
OH O
Glyceraldehyde-3-P
Now have 2 G3P's
CH2
PO3=
Fructose-1,6bisP
OH
+
H
C
C
H
O
CH2
O
PO3=
Dihydroxyacetone-P
Triose isomerase
Glycolytic Pathway for Glucose Metabolism
= OP
3
Triosephosphate
dehydrogenase
NAD+
CH2
O
H
H
C
C
OH O
NADH
{
= OP
3
PO4=
Phosphoglycerate
kinase
O
CH2
H
O
C
C
}
S-Enz
OH
= OP
O
3
ADP
ATP
= OP
3
Glyceraldehyde-3-P
(Metabolism of only
one G3P is shown)
CH2
H
O
C
C
O-PO3=
OH
O
CH2
H
O
C
C
OH
1,3-bisphosphoglycerate
3-phosphoglycerate
OH
Phosphoglycerate
mutase
HO-CH2
H
O
C
C
OH
2-phosphoglycerate
O-PO3=
Enolase
H2O
O
CH2
Pyruvate
kinase
C
C
OH
phosphoenolpyruvate
O-PO3=
ADP
O
ATP
CH3
C
C
O
From the 1 and 6 carbons
of glucose
OH
pyruvate
From the 3 and 4 carbons
of glucose
Isomerization Reaction:
Creates an electron attracting keto group at the # 2 carbon
Mechanism of the isomeration reaction
H
HC
H
C
O
OH
HC
C
OH
O
H
HOCH
HOCH
HCOH
HCOH
HCOH
HCOH
H2CO
H
PO3=
Glucose-6-P
H2CO
C
OH
C
O
HOCH
HCOH
HCOH
PO3=
cis-enolate
H2CO
• H dissociates from C2
• 2 electrons shift
to form cis enediol
• H from hydroxyl group
dissociates
• 2 electrons shift
to form keto group.
• Forces electrons in
enol bond to shift to C1.
PO3=
Fructose-6-P
C-C bond cleavage: Aldolase Reaction
Mechanism of the aldolase reaction
Enol formation
PO3=
H2CO
C
C
O
O-
C
H
+
O
H
HCOH
H2CO
H2CO
CHOH
HOCH
HC
PO3=
H2CO
H
C
C
PO3=
O
O
H
O
Dihydroxyacetone-P
HCOH
PO3=
Carbonyl beta
to Carbon with O
H2CO
PO3=
Glyceraldehyde-3-P
H dissociates from C4; 2 electrons shift to form cis enediol
H from hydroxyl group (C4) dissociates
2 electrons shift to form keto group.
Forces electrons in enol bond to shift to C1.
Coenzymes
(cofactors)/Vitamins
Some are bound to enzyme
Some are soluble
Apoenzyme + cofactor give holoenzyme
Metal ion, organic cofactors
Act as co-substrate
Pyruvate + NADH + H+ --> lactate + NAD+
Vitamins:
Portion of cofactor that cell can’t make, must be in
diet
“Vital amine”
Vitamin forms: Niacin
Vitamin Forms
O
COOH
N
Nicotinic acid
(niacin)
C
NH2
N
Nicotinamide
Nutritional disease: pellagra
Nicotinamide Adenine Dinucleotide
O
C
+
O
HO
P
N
O
O
HO
NH2
CH2 O
H H
H
H
OH OH
NH2
N
P
O
O
CH2 O
H H
H OH OH
N
N
N
PO4= in NADP +
Reduced Form
Oxidized Form
H
C
HC
HC
+
N
R
H
O
C
CH
C
+2H
NH2
-2H
H
C
HC
C
HC
CH
N
R
O
C
NH2
NAD functions
Function: oxidation reduction reaction,
accepts hydride anion (H-): one proton
and two electrons
That’s why we write NADH + H +
Biosynthesis uses NADP+ most often
Catabolism uses NAD+ most often.
In conclusion Streptococcus
Uses glycolysis to degrade glucose to 2
pyruvates
NADH’s made in pathway are reoxidized by
reducing pyruvate to lactate
NADH is key cofactor in oxidation reduction
reactions
ATP made solely by substrate level
phosphorylation.