lecture 3
advertisement
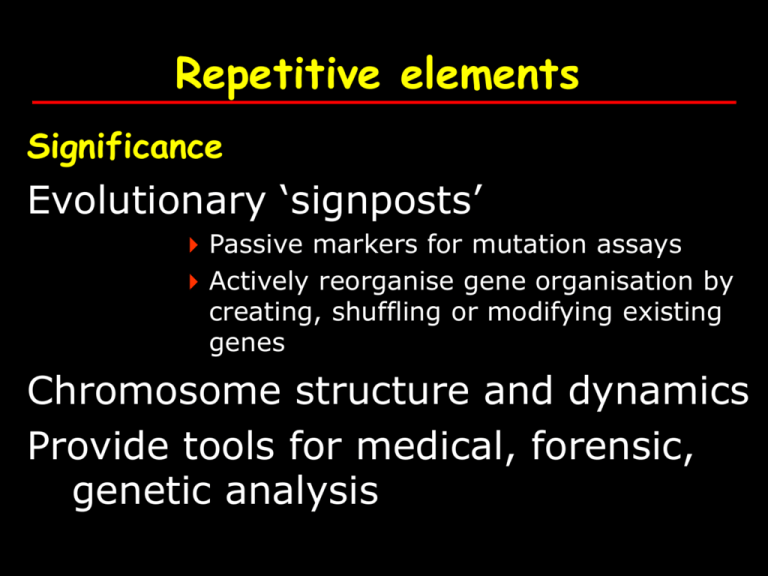
Repetitive elements Significance Evolutionary ‘signposts’ Passive markers for mutation assays Actively reorganise gene organisation by creating, shuffling or modifying existing genes Chromosome structure and dynamics Provide tools for medical, forensic, genetic analysis Repetitive sequences AAA, ATATATAT, CGTCGTCGT etc.. 5 main classes 1) 2) 3) 4) Tandem repeats Transposon-derived repeats Segmental duplications Processed pseudogenes 1) Tandem repeats Blocks of tandem repeats at subtelomeres pericentromeres Short arms of acrocentric chromosomes Ribosomal gene clusters Tandem / clustered repeats Broadly divided into 4 types based on size class Size of repeat Repeat block Satellite 5-171 bp > 100kb Major chromosomal location centromeric heterochromatin minisatellite 9-64 bp 0.1 – 20kb Telomeres microsatellites 1-13 bp < 150 bp Dispersed HMG3 by Strachan and Read pp 265-268 Satellites Large arrays of repeats Some examples Satellite 1,2 & 3 - found in all chromosomes a (Alphoid DNA) b satellite HMG3 by Strachan and Read pp 265-268 Minisatellites Moderate sized arrays of repeats Some examples Hypervariable minisatellite DNA - core of GGGCAGGAXG - found in telomeric regions - used in original DNA fingerprinting technique by Alec Jeffreys HMG3 by Strachan and Read pp 265-268 Microsatellites VNTRs - variable number of tandem repeats, SSR - simple sequence repeats 1-13 bp repeats e.g. (A)n ; (AC)n 2% of genome (dinucleotides - 0.5%) Used as genetic markers (especially for disease mapping) Individual genotype HMG3 by Strachan and Read pp 265-268 Microsatellite genotyping The most common way to detect microsatellites is to design PCR primers that are unique to one locus in the genome and that base pair on either side of the repeated portion Therefore, a single pair of PCR primers will work for every individual in the species and produce different sized products for each of the different length microsatellites Fig 7.7 HMG3 by Strachan and Read pp 190 Microsatellite genotyping . CA repeat genotyping . Marker D17S800 A B C D E Allele types A (3,6) B (1,5) C (3,5) D (2,5) E (3,6) N.B. ‘stutters’ or shadow bands Caused by strand slippage Fig 7.8 HMG3 strand slippage during replication Fig 11.5 HMG3 by Strachan and Read pp 330 strand slippage during replication Fig 11.5 HMG3 by Strachan and Read pp 330 Repetitive elements… 2) Transposon-derived repeats A.k.a. interspersed repeats 45% of genome Arise mainly as a result of transposition either through a DNA or a RNA intermediate 4 main types LINES, SINES, LTRs and DNA transposons Transposon-derived repeats… LINEs (long interspersed elements) Most ancient of eukaryotic genomes Autonomous transposition (reverse trancriptase) ~6-8kb long Internal polymerase II promoter and 2 ORFs 3 related LINE families in humans – LINE-1, LINE-2, LINE-3. Believed to be responsible for retrotransposition of SINEs and creation of processed pseudogenes Nature (2001) pp879-880 HMG3 by Strachan & Read pp268-272 Transposon-derived repeats… SINEs (short interspersed elements) Non-autonomous (successful freeloaders! ‘borrow’ RT from other sources such as LINEs) ~100-300bp long Internal polymerase III promoter No proteins Share 3’ ends with LINEs 3 related SINE families in humans – active Alu, inactive MIR and Ther2/MIR3. Nature (2001) pp879-880 HMG3 by Strachan & Read pp268-272 LINES and SINEs have preferred insertion sites • In this example, yellow represents the distribution of mys (a type of LINE) over a mouse genome where chromosomes are orange. There are more mys inserted in the sex (X) chromosomes. Try the link below to do an online experiment which shows how an Alu insertion polymorphism has been used as a tool to reconstruct the human lineage http://www.geneticorigins.org/geneticorigins/ pv92/intro.html Transposon-derived repeats… Long Terminal Repeats (LTR) Repeats on the same orientation on both sides of element e.g. ATATATNNNNNNNATATAT Autonomous or non-autonomous Autonomous retroposons encode gag, pol genes which encode the protease, reverse transcriptase, RNAseH and integrase Nature (2001) pp879-880 HMG3 by Strachan & Read pp268-272 Transposon-derived repeats… DNA transposons (lateral transfer?) DNA transposons Inverted repeats on both sides of element e.g. ATGCNNNNNNNNNNNCGTA From GenesVII by Levin Nature (2001) pp879-880 Transposon derived repeats major types class family size Copies* LINE LINE-1 (Kpn family) ~6.4kb 0.8x106 % genome* 15.4 SINE Alu ~0.3kb 1.3x106 10.7 LTR e.g.HERV ~1.3kb 0.7x106 7.9 ~0.25kb 0.4x106 2.7 DNA transposon mariner * Updated from HGP publications HMG3 by Strachan & Read pp268-272 3) Segmental duplications Closely related sequence blocks at different genomic loci Transfer of 1-200kb blocks of genomic sequence Segmental duplications can occur on homologous chromosomes (intrachromosomal) or non homologous chromosomes (interchromosomal) Not always tandemly arranged Relatively recent Segmental duplications Interchromosomal segments duplicated among non-homologous chromosomes Intrachromosomal duplications occur within a chromosome / arm Nature Reviews Genetics 2, 791-800 (2001); Segmental duplications in chromosome Segmental 22 duplications Segmental duplications - chromosome 7. Nature Reviews Genetics 2, 791-800 (2001) 4) Pseudogenes - processed Repetitive sequences AAA, ATATATAT, CGTCGTCGT etc.. 5 main classes 1) Tandem repeats 2) Transposon-derived repeats 3) Segmental duplications 4) Processed pseudogenes Insights from the HGP……… 7) Repeat content a) Age distribution b) Comparison with other genomes c) Variation in distribution of repeats d) Distribution by GC content e) Y chromosome Nature (2001) 409: pp 879-891 Repeat content……. a) Age distribution Most interspersed repeats predate eutherian radiation (confirms the slow rate of clearance of nonfunctional sequence from vertebrate genomes) LINEs and SINEs have extremely long lives 2 major peaks of transposon activity No DNA transposition in the past 50MYr LTR retroposons teetering on the brink of extinction a) Age distribution overall decline in interspersed repeat activity in hominid lineage in the past 35-40MYr compared to mouse genome, which shows a younger and more dynamic genome b) Comparison with other genomes Higher density of transposable elements in euchromatic portion of genome Higher abundance of ancient transposons 60% of IR made up of LINE1 and Alu repeats whereas DNA transposons represent only 6% (a few human genes appear likely to have resulted from horizontal transfer from bacteria!!) c) Variation in distribution of repeats Some regions show either High repeat density e.g. chromosome Xp11 – a 525kb region shows 89% repeat density Low repeat density e.g. HOX homeobox gene cluster (<2% repeats) (indicative of regulatory elements which have low tolerance for insertions) d) Distribution by GC content High GC – gene rich ; High AT – gene poor LINEs abundant in AT-rich regions SINEs lower in AT-rich regions Alu repeats in particular retained in actively transcribed GC rich regions E.g. chromosme 19 has 5% Alus compared to Y chromosome Repeat content……. e) The Y chromosome ! Unusually young genome (high tolerance to gaining insertions) Mutation rate is 2.1X higher in male germline Possibly due to cell division rates or different repair mechanisms • Working draft published – Feb 2001 • Finished sequence – April 2003 • Annotation of genes going on References Text: 1) Human Molecular Genetics 3 by Strachan and Read – Chapter 9 pp 265-268 Optional Reading 1) 2) Batzer MA, Deininger PL Alu repeats and human genomic diversity Nature Rev Genet 3 (5): 370-379 May 2002 BS Emanuel & TH Shaikh Segmental duplications: an 'expanding' role in genomic instability and disease Nature Reviews Genetics 2, 791-800 (2001) 3) Nature (2001) 409: pp 879-891