Biology 4 Lecture - Eukaryotic Genomes (Ch 19)
advertisement
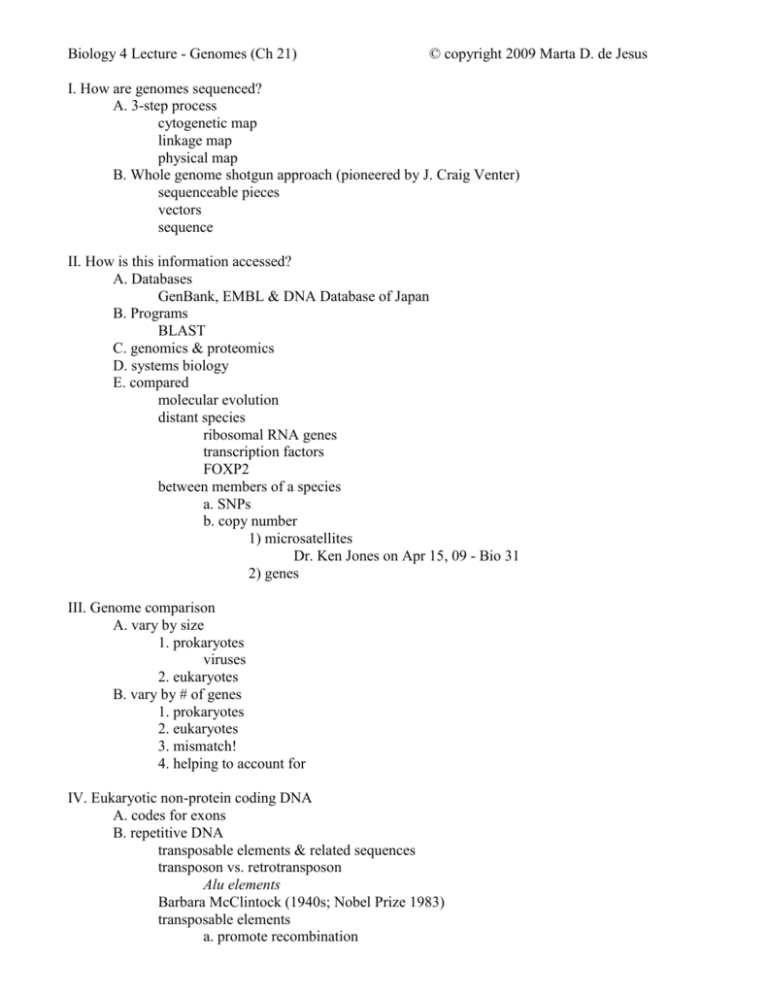
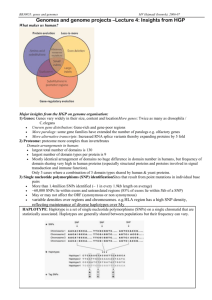
Biology 4 Lecture - Genomes (Ch 21) © copyright 2009 Marta D. de Jesus I. How are genomes sequenced? A. 3-step process cytogenetic map linkage map physical map B. Whole genome shotgun approach (pioneered by J. Craig Venter) sequenceable pieces vectors sequence II. How is this information accessed? A. Databases GenBank, EMBL & DNA Database of Japan B. Programs BLAST C. genomics & proteomics D. systems biology E. compared molecular evolution distant species ribosomal RNA genes transcription factors FOXP2 between members of a species a. SNPs b. copy number 1) microsatellites Dr. Ken Jones on Apr 15, 09 - Bio 31 2) genes III. Genome comparison A. vary by size 1. prokaryotes viruses 2. eukaryotes B. vary by # of genes 1. prokaryotes 2. eukaryotes 3. mismatch! 4. helping to account for IV. Eukaryotic non-protein coding DNA A. codes for exons B. repetitive DNA transposable elements & related sequences transposon vs. retrotransposon Alu elements Barbara McClintock (1940s; Nobel Prize 1983) transposable elements a. promote recombination b. change c. modify d. change 6. other repetitive DNA a. large-segment duplication b. simple sequence http://www.vivo.colostate.edu/hbooks/genetics/medgen/dnatesting/satellites.html (edited) 1) satellites are very highly repetitive (i) repeat lengths of one to several thousand base pairs. (ii) typically are organized as large (up to 100 million bp !) clusters in the heterochromatic regions of chromosomes, near centrosomes and telomeres (iii) these are also found abundantly on the Y chromosome. 2) minisatellites are moderately repetitive (i) tandemly repeated arrays of moderately-sized (9 to 100 bp, but usually about 15 bp) repeats (ii) generally involving mean array lengths of 0.5 to 30 kb. (iii) found in euchromatic regions of the genome of vertebrates, fungi and plants and are highly variable in array size. (iv) most highly polymorphic sequence elements yet discovered in the human genome, and delineating the repeat lengths of these loci is the basis of most DNA typing systems used in forensic medicine. 3) microsatellites = simple sequence repeats (SSR) = short tandem repeats (STR) = variable number tandem repeats (VNTR) are moderately repetitive: (i) arrays of short (2-6 bp) repeats found in vertebrate, insect and plant genomes of arrays 10-100 bp long. (ii) classified according to length as (mono-), di-, tri-, tetra-, penta-, and hexanucleotide repeats. (iii) human genome contains at least 30,000 microsatellite loci located in euchromatin. (iv) copy numbers are characteristically variable within a population so used for population genetics studies (forensics, paternity analysis, wildlife management, etc.) 4) In general, satellite DNAs show exceptional variability among individuals, particularly with regard to the number of repeats at a given locus. C. other non-coding DNA 1. introns & regulatory sequences 2. unique non-coding DNA V. Genes multigene families 1. eg: ribosomal RNAs 2. eg: - & - globin genes contributing to genome evolution 1. mutation 2. duplications 3. multiple copies pseudogenes 4. exon shuffling D. comparing in development: Evo-Devo 1. homeotic genes 2. in metazoans: Hox genes homeobox homeodomain 3. in plants flower development major master set is Mads-box