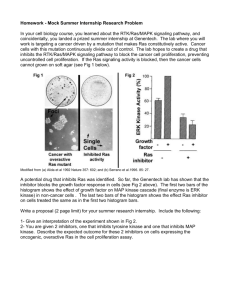
Document
advertisement

MCB Cell Signaling Lectures 1 & 2 Ken Blumer Dept. of Cell Biology & Physiology 506 McDonnell Sciences kblumer@wustl.edu 362-1668 Lecture 1 General Concepts of Signal Transduction Cell Communication Types of Receptors Molecular Signaling Receptor Binding Scatchard Analysis Competitive Binding Second Messengers G proteins Modes of cell communication Lodish, 20-1 Four classes of cell-surface receptors Lodish, 20-3 Transmitting/transducing signals within cells: 3 basic modes (may be combined) 1. Allostery Shape change, often induced by binding a protein or small molecule Switching can be very rapid P 2. Covalent modification Modification itself changes molecule’s shape Memory device; may be reversible (or not) 3. Proximity (= regulated recruitment) Regulated molecule may already be in “signaling mode;” induced proximity to a target promotes transmission of the signal P P Finding and analyzing receptors: Ligand binding assays Receptor: ligand binding must be specific, saturable, and of high affinity Saturation Binding studies Can be performed in intact cells, membranes, or purified receptors 1. Add various amounts of labeled ligand (drug, hormone, growth factor) 2. To determine specific binding, add an excess of unlabeled ligand to compete for specific binding sites. QU: Why is there non-specific binding? 3. Bind until at equilibrium 4. Separate bound from unbound ligand 5. Count labeled ligand [Adapted from A. Ciechanover et al., 1983, Cell 32:267.] Receptor abundance, affinity, cooperativity: Scatchard plots (Bound Lig) (Free) Slope = - 1/Kd X intercept = # rec (Bound Lig) For an excellent discussion of principles of receptor binding, and practical considerations, see http://www.graphpad.com; also posted on MCB website. Cooperativity indicated by non-linear Scatchard plots Positive cooperativity: binding of ligand to first subunit increases Affinity of subsequent binding events. Example: hemoglobin binding O2 (Bound Lig) (Free) (Bound Lig) Negative cooperativity: binding of ligand to first subunit decreases affinity of subsequent binding events. What receptors do: Generate second messengers Molecular mediators of signal transduction. Cells carefully, and rapidly, regulate the intracellular concentrations. Second messengers can be used by multiple signaling networks (at the same time). • • • • • • Cyclic nucleotides: cAMP, cGMP Inositol phosphate (IP) Diacylglycerol (DAG) Calcium Nitric oxide (NO) Reactive oxygen species (ROS) cAMP regulates protein kinase (PKA) activity Positive cooperativity--binding of increases affinity for second cAMP PKA targets include Phosphorylase kinase and the transcription regulator, cAMP response element binding (CREB) protein Alberts 15-31,32 Lipid-derived second messengers: Diacylglycerol and inositol phosphates Alberts, 15-35 IP3 evokes calcium release as third messenger Lodish, 20-39 A key effector of Ca2+-CaM: CaM-kinase II Alberts, 15-41 NO signaling Gases can act as second messengers! NO effects are local, since it has half-life of 5-10 seconds (paracrine). NO activates guanylate cyclase by binding heme ring (allosteric mechanism) Lodish, 20-42 Discovery of NO signaling Furchgott, Ignarro, Murad, Nobel Prize 1998 Robert F Furchgott showed that acetylcholine-induced relaxation of blood vessels was dependent on the endothelium. His "sandwich" experiment set the stage for future scientific development. He used two different pieces of the aorta; one had the endothelial layer intact, in the other it had been removed. Louis Ignarro reported that EDRF relaxed blood vessels. He also identified EDRF as a molecule by using spectral analysis of hemoglobin. When hemoglobin was exposed to EDRF, maximum absorbance moved to a new wave-length; and exposed to NO, exactly the same shift in absorbance occurred! EDRF was identical with NO. http://www.nobel.se/medicine/laureates/1998/illpres/index.html G proteins: Switches linking receptors & 2nd messengers • Discovery and Structure of Heterotrimeric G proteins • Signaling pathways of G proteins • Receptors that activate G proteins • Small G proteins-discovery and structure • Activation and inactivation mechanisms • Alliance for Cell Signaling (AfCS) Signal Transduction by G proteins • Discovery and Structure of Heterotrimeric G proteins • Signaling pathways of G proteins • Receptors that activate G proteins • Small G proteins-discovery and structure • Activation and inactivation mechanisms • Alliance for Cell Signaling (AfCS) G protein signal transduction Neves, Ram, Iyengar, Science 2002 Hydrolysis of GTP • • • • Arg & Gln stabilize the b and g phospates of GTP molecule in correct orientation for hydrolysis by H2O Hydrolysis leads to major conformation change in Gs a Mutations in the Gln or Arg (or ADP ribosylation by cholera toxin) blocks the ability to stabilize transition state, and therefore locks G protein in the “on” position. Examples include adenomas of pituitary and thyroid glands (GH secreting tumors, acromegaly), and McCune-Albright syndrome. Iiri, et al. NEJM (1999) Signal Transduction by G proteins • Discovery and Structure of Heterotrimeric G proteins • Signaling pathways of G proteins • Receptors that activate G proteins • Small G proteins-discovery and structure • Activation and inactivation mechanisms • Alliance for Cell Signaling (AfCS) G protein-coupled receptors (GPCRs) • • • • Many ligands Robust switches Multiple effectors Conserved 7 TM architecture • More than 50% of drugs target GPCRs Lefkowitz Kobilka Bockaert & Pin, EMBO J (1999) 2012 Nobel Prize GPCR desensitization mechanisms Arrestins act as scaffolds for ERK and JNK signaling pathways Lefkowitz reviews Signal Transduction by G proteins • Discovery and Structure of Heterotrimeric G proteins • Signaling pathways of G proteins • Receptors that activate G proteins • Small G proteins-discovery and structure • Activation and inactivation mechanisms • Alliance for Cell Signaling (AfCS) Reverse genetics: express one or two mutant versions of the protein of interest Depends on understanding how the machines work 1. Inhibit activity of the protein with a “dominant-negative” interfering mutant of that protein The mutant titrates (binds up) a limiting component to block the normal protein’s signal 2. Increase activity of the protein with a “dominant-positive” or “constitutively active” interfering mutant of the protein The mutant exerts the same effect as the normal protein would, if it were activated in the cell Reverse genetics: small GTPases as examples Depends on understanding how the machines work “Dominant-negative” mutation GEF “Dominant-positive” mutation GDP Binds GEF but cannot replace GDP by GTP; so GEF not available for activating normal protein GEF GDP GTP Pi The mutant titrates (binds up) a limiting component to block the normal protein’s signal Cannot hydrolyze GTP, so remains always active GAP The mutant exerts the same effect as the normal protein would, if it were activated Signal Transduction by G proteins • Discovery and Structure of Heterotrimeric G proteins • Signaling pathways of G proteins • Receptors that activate G proteins • Small G proteins-discovery and structure • Activation and inactivation mechanisms Small G protein “turn on” mechanisms First mammalian GEF, Dbl, isolated in 1985 as an oncogene in NIH 3T3 focus forming assay. It had an 180 amino acid domain with homology to yeast CDC24. This domain, named DH (Dbl homology) is necessary for GEF activity. In 1991, Dbl shown to catalyze nucleotide exchange on Cdc42. Schmidt & Hall, Genes & Dev. (2002) Dbl= Diffuse B-cell lymphoma Many RhoGAPs RhoGAPs outnumber the small G proteins Rho/Rac/Cdc42 by nearly 5fold. Why so much redundancy? Luo group did RNAi against 17 of the 20 RhoGAPs in fly. Six caused lethality when expressed ubiquitously. Tissue specific expression of RNAi revealed unique phenotypes. P190RhoGAP implicated in axon withdrawal. Increasing amounts of RNAi caused more axon withdrawal (panels C-G). Why so many RhoGAPs? Billuart, et al. Cell (2001) The GTPase switch Schmidt & Hall, Genes & Dev. (2002) Growth Factors and Receptor Tyrosine Kinases • • • • • RTK’s--How do they work? EGFR signaling and ras MAP kinase cascades PI3K, PKB, PLCg PTPs (Protein Tyrosine Phosphatases) How RTKs (& TK-linked Rs) work 1. Ligand promotes formation of RTK dimers, by different mechanisms: Ligand itself is a dimer (PDGF) One ligand binds both monomers (GH) 2. Dimerization allows trans-phosphorylation of catalytic domains, which induces activation of catalytic (Y-kinase) activity 3. Activated TK domains phosphorylate each other and proteins nearby, sometimes on multiple tyrosines 4. Y~P residues recruit other signaling proteins, generate multiple signals EGF receptor as a model 1st RTK to be characterized v-erbB oncogene = truncated EGFR How do we know that the EGFR autophosphorylates in trans? Experiment: test WT and short EGFRs, each with or without a kin- mutation wt kinshort kinshort kin+ + + + + + + Honneger et al. (in vitro) PNAS 1989; (in vivo) MCB 1999 Does this result rule out phosphorylation in cis as well? If not, how can you find out? PS: What do trans and cis mean? How can we know that the EGFR does not autophosphorylate in cis? Need an EGFR that cannot homodimerize EGFR family is huge, with many RTK members and many EGF-like ligands Such receptors often form obligatory heterodimers with a similar but different partner If A can dimerize only with A’, then we can inactivate the kinase domain of A’ and ask whether A phosphorylates itself Answer: NO QED Growth Factors and Receptor Tyrosine Kinases • • • • • RTK’s--How do they work? EGFR signaling and ras MAP Kinase Cascades PI3K, PKB, PLCg PTPs (Protein Tyrosine Phosphatases) Signals generated by the EGFR Individual Y~P residues recruit specific proteins, generate different signals The activated dimer phosphorylates itself P . SOS, a Ras GEF . P T-loop only P . P P . Multiple sites P Docks via intermediate adapters to activate Ras Ras activates multiple targets (MAPK) PLC-g Docking of Y-kinases allows Tyr-phos’n of PLC-g, which activates it PI3-kinase Adapters again Docking allosterically activates PI3K Each signal, in turn, activates a different set of pathways, which cooperate to produce the overall response P P P Adapters connect A with B, B with C . . . to create complex, localized assemblies of signaling proteins Adapter 2 Each adapter has at least 2 interaction domains, and may have other functions as well Types of adapter interactions A B P C Adapter 1 Y~P sequence motifs allow regulatable adapter functions Also SH2 PTB Tyrosine phosphates Tyrosine phosphates SH3 Polyproline-containing sequences PDZ Pleckstrin homol. (PH) Many others Specific 4-residue sequences at C-termini Phosphoinositides EGF activates the MAPK pathway in multiple steps, with multiple mechanisms EGF Extracellular GF EGFR RTK EGFR~P Phospho-RTK Grb2 Adapter SOS Ras Small GTPase Mechanism Proximity Allostery Covalent modification Ras-GEF Raf Ser kinase Tyr/thr kinase Mek Ser kinase Transcription factor ERKs C-Jun EGFR Activation of Ras: Proximity & Allostery The Players RTK = EGFR P . P P . P P Ras GDP P “GF receptor binding 2” Adapter, found in screen for binders to EGFR~P SH3 SH2 Grb2 SH3 SOS “Rat Sarcoma” Small GTPase, attached to PM by prenyl group “Son of Sevenless” GEF, converts Ras-GDP to Ras-GTP Found in Drosophila, homol. To S.c. Cdc25 EGFR Activation of Ras: Proximity & Allostery Even before EGF arrives . . . . . Ras GDP SOS is “ready to go”: already (mostly) associated with Grb2 in cytoplasm, in the resting state SH3 SH2 Grb2 SH3 SOS EGFR Activation of Ras: Proximity & Allostery Then . . . Covalent modification P . P P . P P Ras GDP P EGF-bound dimers trigger phosphorylation, in trans SH3 SH2 Grb2 SH3 SOS EGFR Activation of Ras: Proximity & Allostery Then . . . Proximity P . P P . P P P SH2 Ras GDP SH3 Grb2 SOS SH3 Grb2’s SH2 domain binds Y~P on EGFR, bringing SOS to the plasma membrane EGFR Activation of Ras: Proximity & Allostery Then . . . Allostery P . P P . P P P SH2 Ras GDP SH3 Grb2 SOS SH3 GDP SOS now binds Ras-GDP, causing GDP to dissociate, and . . . EGFR Activation of Ras: Proximity & Allostery Then . . . Allostery continues P . P P . P P P SH2 Ras GTP SH3 Grb2 SOS SH3 GTP GTP enters empty pocket on Ras, which dissociates from SOS and converts into its active conformation EGFR Activation of Ras: Proximity & Allostery Finally . . . Proximity again! P . P P . P P P SH2 Ras GTP SH3 Grb2 Raf SOS SH3 GTP Ras-GTP brings Raf to the PM for activation, and the MAPK cascade is initiated Raf MAPK Cascade Growth Factors and Receptor Tyrosine Kinases • • • • • RTK’s--How do they work? EGFR signaling and ras MAP Kinase Cascades PI3K, PKB, PLCg PTPs (Protein Tyrosine Phosphatases) The best understood MAPK cascade MAPK = Mitogen-activated protein kinase . Raf-1 A-raf B-raf MAPKKK Phos’n of T-loop Ser residues . P P Phos’n of T-loop Thr and Tyr MEK1 MEK2 . MAPKK P P Phos’n of Ser/Thr ERK1 ERK2 MAPK C-Jun Altered gene expression Switch-like behavior* Responses are not always graded 1.0 Progesterone MAPKKK Response Instead . . . Frog oocyte MAPKK 0.5 0 MAPK 0 1 5 Stimulus (multiples of EC50) G2-M transition Amplified sensitivity: reduces noise @ low stimulus; reversible Bistable responses: off or on, often via positive feedback & used for irreversible responses (e.g., cell cycle) Other examples? *JE Ferrell, Tr Bioch Sci 22:288, 1997 All or nothing response in Xenopus oocytes Progesterone, or fertilization, induces germinal vesicle breakdown of Xenopus oocytes--a process mediated by the MAPK cascade. Question: At a concentration of progesterone that halfmaximally activates MAPK (0.01 uM, panel A), are all the oocytes activated halfway (panel B), or are half of the oocytes activated fully (panel C)? Since Xenopus oocytes are HUGE, one can look at MAPK on a cell by cell basis. Ferrell, et al., Science (1998) Answer: All or nothing. Scaffold proteins involved in ERK-signaling pathways Dhanasekaran (2007) Oncogene Growth Factors and Receptor Tyrosine Kinases • • • • • RTK’s--How do they work? EGFR signaling and ras MAP Kinase Cascades PI3K, PKB, PLCg PTPs (Protein Tyrosine Phosphatases EGFR Activation of PI3K combines Proximity & Allostery P . . PIP2 P SH2 P P Activated by EGFR/p85 Can also be activated by Rac or Ras! PIP3 SH2 p85 p110 SH2 Recruitment from cytoplasm to PM, via SH2 domains SH2 p85 p110 How do we know proximity is not enough? 1. p85 mutants that activate without binding to RTKs 2. Tethering to membrane does not activate PIP3 targets include many GEFs, many tyrosine kinases, and others, including . . . PKB (aka Akt) = ser/thr kinase that promotes cell survival P P P P PIP3 PKB (= membrane lipid) PH K . . . is inactive in cytoplasm . . . contains a PH (pleckstrin homology) domain & a kinase domain Multi-step activation of PKB: proximity PIP3 P P PH P K PH domain recognizes 3’phosphate of PIP3, bringing kinase domain to the PM P Proximity to PM alone does not activate the kinase PH K Multi-step activation of PKB: covalent modification PIP3 P P PH P P Inactive PKB PDK1* K P P PH P P K P P Active (phos’d) PKB *PDK1 is also recruited to the membrane via a PIP3-binding PH domain Overall, two proximity steps plus (at least) one phosphorylation step EGFR Activation of PLCg combines THREE inputs P . . P P P PIP3 P P P P PIP2 P P PLCg (Inactive, in cytoplasm) PH SH2 1. PROXIMITY: Recruitment from cytoplasm to PM, via SH2 domains SH2 Catalytic EGFR Activation of PLCg combines THREE inputs 3. PROXIMITY: Binds to PIP3 via PH domain P . . P P P P P P P PIP2 DAG P SH2 P 2. COVALENT: Activated by EGFR phosph’n PH SH2 P Catalytic InsP3 Growth Factors and Receptor Tyrosine Kinases • • • • • RTK’s--How do they work? EGFR signaling and ras MAP Kinase Cascades PI3K, PKB, PLCg PTPs (Protein Tyrosine Phosphatases) PTEN opposes PI3K by removing PI3-phosphate PTEN discovered as a tumor suppressor gene. Mutated in brain, breast and prostate cancers. Has homology to dual specificity phosphates, but shows little activity toward phosphoproteins. Was discovered to remove phosphates from PIPs; thereby providing likely mechanism for tumor suppression. Cantley & Neel, PNAS (1999) Gleevec--proof that you can target kinases for drug therapy