EGFR Activation of Ras
advertisement

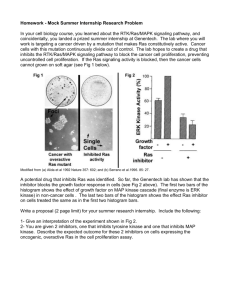
Growth Factors and Receptor Tyrosine Kinases • • • • • RTK’s--How do they work? EGFR signaling and ras MAP kinase cascades PI3K, PKB, PLCg PTPs (Protein Tyrosine Phosphatases) Epidermal growth factor Neurotrophic growth factor (NGF) isolated from mouse submaxillary glands (Rita Levi-Montalcini) Stanley Cohen, 1962 “Side effects” of impure NGF preps Premature eyelid opening (7d vs. 14 d) Premature tooth eruption (6 d vs. 9 d) Pure “Tooth-lid factor” = EGF Important roles in development no EGF EGF 1 mg/kg Mitogenic for fibroblasts Regulates growth/differentiation of many target cells Refs: S. Cohen, JBC 237:1555, 1962 S. Cohen, Nobel lecture, 1986 Phospho-tyrosine signals Kinases phosphorylate tyrosine (Y*) residues of target proteins Y~P = target for distinctive protein binding pockets, with surrounding sequences lending specificity ALWAYS activate by promoting proximity of proteins A and B (sometimes by allostery also) A X TK A P X A P B X In its new proximity to A, B’s activity (= X) can now: Phosphorylate or de-phosphorylate another protein Make or degrade a 2nd messenger Attract additional signaling molecules B Y~P provides long-lasting but erasable memory, which is terminated by DE-phosphorylation *Y = one-letter code for tyrosine; S = ser, T = thr, etc. Phospho-tyrosine signals regulate growth & differentiation RTKs = Receptor Tyrosine Kinases Extracellular region variable, with many different motifs Usually cross membrane only once Intracellular region contains conserved catalytic domains ALSO: TK-linked receptors for: Antigens (receptors on B and T cells Growth hormone Interleukin-4 Erythropoietin, many others Alberts, 15-47 How RTKs (& TK-linked Rs) work 1. Ligand promotes formation of RTK dimers, by different mechanisms: Ligand itself is a dimer (PDGF) One ligand binds both monomers (GH) 2. Dimerization allows trans-phosphorylation of catalytic domains, which induces activation of catalytic (Y-kinase) activity 3. Activated TK domains phosphorylate each other and proteins nearby, sometimes on multiple tyrosines 4. Y~P residues recruit other signaling proteins, generate multiple signals EGF receptor as a model 1st RTK to be characterized v-erbB oncogene = truncated EGFR Evidence for EGFR dimerization Yarden & Schlessinger Rate of phosphorylation = k[EGFR]2, even in micelles! Therefore: 2 EGFRs required for phosphorylation Later confirmed by Chemical cross-linking FRET Dominant-negative mutants (e.g., kinase-dead EGFR) IMPORTANT Dimerization/proximity = alternative to allostery (Shown by swapping EC/IC domains of EGFR, PDGFR) How do we know that the EGFR autophosphorylates in trans? Experiment: test WT and short EGFRs, each with or without a kin- mutation wt kinshort kinshort kin+ + + + + + + Honneger et al. (in vitro) PNAS 1989; (in vivo) MCB 1999 Does this result rule out phosphorylation in cis as well? If not, how can you find out? PS: What do trans and cis mean? How can we know that the EGFR does not autophosphorylate in cis? Need an EGFR that cannot homodimerize EGFR family is huge, with many RTK members and many EGF-like ligands Such receptors often form obligatory heterodimers with a similar but different partner If A can dimerize only with A’, then we can inactivate the kinase domain of A’ and ask whether A phosphorylates itself Answer: NO QED How does dimerization activate RTKs? GFRs (like many kinases) have sites in their T loops at which phosphorylation activates Dimerization induces T-loop phosphorylation in trans Phosphorylation of Y (one or more) in T-loop causes it to move out of the way of the active site. T-loop Cat. loop Y1162 occupies the active site Substrate Y sits in active site Proximity by itself is usually enough to promote T-loop phosphorylation, but there may also be a role for allostery Once activated, each monomer can phosphorylate nearby Y residues in the other, as well as in other proteins Y1162 flips out Growth Factors and Receptor Tyrosine Kinases • • • • • RTK’s--How do they work? EGFR signaling and ras MAP Kinase Cascades PI3K, PKB, PLCg PTPs (Protein Tyrosine Phosphatases) Signals generated by the EGFR Individual Y~P residues recruit specific proteins, generate different signals The activated dimer phosphorylates itself P . SOS, a Ras GEF . P T-loop only P . P P . Multiple sites P Docks via intermediate adapters to activate Ras Ras activates multiple targets (MAPK) PLC-g Docking of Y-kinases allows Tyr-phos’n of PLC-g, which activates it PI3-kinase Adapters again Docking allosterically activates PI3K Each signal, in turn, activates a different set of pathways, which cooperate to produce the overall response P P P Adapters connect A with B, B with C . . . to create complex, localized assemblies of signaling proteins Adapter 2 Each adapter has at least 2 interaction domains, and may have other functions as well Types of adapter interactions A B P C Adapter 1 Y~P sequence motifs allow regulatable adapter functions Also SH2 PTB Tyrosine phosphates Tyrosine phosphates SH3 Polyproline-containing sequences PDZ Pleckstrin homol. (PH) Many others Specific 4-residue sequences at C-termini Phosphoinositides SH2 & SH3 domains--src homology domains SH domains are protein domains initially discovered in Src, a transforming tyrosine kinase found in Rous sarcoma virus. Sequences of many signaling proteins that interact with RTKs revealed multiple homologous domains to Src region 2 and region 3. Lodish, 24-17 SH2: Protein motif of ~100 amino acids, binds to phosphotyrosine peptide sequences. (87 SH2 in the human genome) SH3: ~60 amino acid domain, binds to R-X-X-P-X-X-P peptide sequences. (143 SH3 in the human genome) How would you determine the specificity of an individual SH2 domain for a phosphopeptide? EGF activates the MAPK pathway in multiple steps, with multiple mechanisms EGF Extracellular GF EGFR RTK EGFR~P Phospho-RTK Grb2 Adapter SOS Ras Small GTPase Mechanism Proximity Allostery Covalent modification Ras-GEF Raf Ser kinase Tyr/thr kinase Mek Ser kinase Transcription factor ERKs C-Jun Fly genetics to the rescue Fly eye consists of ~800 ommatidia, an individual lens structure consisting of 22 cells (8 photoreceptor cells, R1-R8) Eye development is highly ordered process. RTK signaling is essential. Mutation in sevenless results in loss of R7. Additional mutations in pathway identified sos (son-of-sevenless), boss (bride of sevenless), Drk (downstream of receptor kinase) Alberts, 15-53 EGFR Activation of Ras: Proximity & Allostery The Players RTK = EGFR P . P P . P P Ras GDP P “GF receptor binding 2” Adapter, found in screen for binders to EGFR~P SH3 SH2 Grb2 SH3 SOS “Rat Sarcoma” Small GTPase, attached to PM by prenyl group “Son of Sevenless” GEF, converts Ras-GDP to Ras-GTP Found in Drosophila, homol. To S.c. Cdc25 EGFR Activation of Ras: Proximity & Allostery Even before EGF arrives . . . . . Ras GDP SOS is “ready to go”: already (mostly) associated with Grb2 in cytoplasm, in the resting state SH3 SH2 Grb2 SH3 SOS EGFR Activation of Ras: Proximity & Allostery Then . . . Covalent modification P . P P . P P Ras GDP P EGF-bound dimers trigger phosphorylation, in trans SH3 SH2 Grb2 SH3 SOS EGFR Activation of Ras: Proximity & Allostery Then . . . Proximity P . P P . P P P SH2 Ras GDP SH3 Grb2 SOS SH3 Grb2’s SH2 domain binds Y~P on EGFR, bringing SOS to the plasma membrane EGFR Activation of Ras: Proximity & Allostery Then . . . Allostery P . P P . P P P SH2 Ras GDP SH3 Grb2 SOS SH3 GDP SOS now binds Ras-GDP, causing GDP to dissociate, and . . . EGFR Activation of Ras: Proximity & Allostery Then . . . Allostery continues P . P P . P P P SH2 Ras GTP SH3 Grb2 SOS SH3 GTP GTP enters empty pocket on Ras, which dissociates from SOS and converts into its active conformation EGFR Activation of Ras: Proximity & Allostery Finally . . . Proximity again! P . P P . P P P SH2 Ras GTP SH3 Grb2 Raf SOS SH3 GTP Ras-GTP brings Raf to the PM for activation, and the MAPK cascade is initiated Raf MAPK Cascade How does Ras activate Raf? Proximity vs. allostery? Allostery: Ras recruits Raf to the PM and activates it directly Ras Ras GTP GTP Raf* Raf MAPK Cascade (Cytoplasmic) Proximity: Ras recruits Raf to the PM, where it is activated by X X Ras Ras GTP GTP Raf (Cytoplasmic) Raf* MAPK Cascade How can we tell the difference? Does Raf signal (without Ras) when recruited to the PM? Stokoe et al. (1994) Science Experiment Attach a CAAX* box to Raf’s Cterminus Express Raf-CAAx in cells, measure activity of MEK, an enzyme downstream in the MAPK pathway EXV Raf Raf+RasG12 RafCAAX RafCAAX+Ras17N RafCAAX+RasG12V 0 10 20 Relative MEK activity *CAAX (A = aliphatic; C = cysteine) is a site for prenylation; prenylated proteins concentrate at the PM Answer: “proximity +” Ras does localize Raf but does not activate it (other proteins do) Growth Factors and Receptor Tyrosine Kinases • • • • • RTK’s--How do they work? EGFR signaling and ras MAP Kinase Cascades PI3K, PKB, PLCg PTPs (Protein Tyrosine Phosphatases) Mammalian MAP Kinase Cascades Johnson & Lapadat (2002) Science 298: 1911 Borrowed from Chan, STKE The best understood MAPK cascade MAPK = Mitogen-activated protein kinase . Raf-1 A-raf B-raf MAPKKK Phos’n of T-loop Ser residues . P P Phos’n of T-loop Thr and Tyr MEK1 MEK2 . MAPKK P P Phos’n of Ser/Thr ERK1 ERK2 MAPK C-Jun Altered gene expression MAPK “cassettes” mediate many different responses Vertebrates Frog oocyte Mitogens Progesterone MAPKKK MAPKKK MAPKKK MAPKK MAPKK MAPKK MAPK MAPK MAPK G2-M transition Cell cycle arrest, mating Proliferation S. cerevisiae Mating pheromone Additional sites for regulation Different biology, similar cassettes: why 3 kinases? Combinatorial diversity Magnitude amplification Switch-like responses Switch-like behavior* Responses are not always graded 1.0 Progesterone MAPKKK Response Instead . . . Frog oocyte MAPKK 0.5 0 MAPK 0 1 5 Stimulus (multiples of EC50) G2-M transition Amplified sensitivity: reduces noise @ low stimulus; reversible Bistable responses: off or on, often via positive feedback & used for irreversible responses (e.g., cell cycle) Other examples? *JE Ferrell, Tr Bioch Sci 22:288, 1997 All or nothing response in Xenopus oocytes Progesterone, or fertilization, induces germinal vesicle breakdown of Xenopus oocytes--a process mediated by the MAPK cascade. Question: At a concentration of progesterone that halfmaximally activates MAPK (0.01 uM, panel A), are all the oocytes activated halfway (panel B), or are half of the oocytes activated fully (panel C)? Since Xenopus oocytes are HUGE, one can look at MAPK on a cell by cell basis. Ferrell, et al., Science (1998) Answer: All or nothing. Of course, life is not so simple . . . BONUS slide Does this work in mammalian cells? Blenis and co-workers used FACS and immunohistochemistry (anti-DP ERK Ab) to look at EGF activation of ERK in Swiss 3T3 fibroblasts MacKeigan MCB 2005 Scaffolds for MAP Kinase signaling Deletion analysis of the binding of JIP-1 to JNK1, MKK7, MLK3, and DLK. JIP-1 was expressed in cells as a GST fusion protein together with HPK1 or epitope-tagged JNK1, MKK7, MLK3, and DLK (15, 16). The presence of these kinases in glutathione-agarose precipitates was examined by protein immunoblot analysis. HPK=hematopoeitic progenitor kinase DLK=dual lineage kinase (member of the MLK family) Whitmarsh et. al. (1998) Science 281: 1671 Scaffolding roles of JNK-interacting proteins Dhanasekaran (2007) Oncogene Scaffold proteins involved in ERK-signaling pathways Dhanasekaran (2007) Oncogene Growth Factors and Receptor Tyrosine Kinases • • • • • RTK’s--How do they work? EGFR signaling and ras MAP Kinase Cascades PI3K, PKB, PLCg PTPs (Protein Tyrosine Phosphatases EGFR Activation of PI3K combines Proximity & Allostery P . . PIP2 P SH2 P P Activated by EGFR/p85 Can also be activated by Rac or Ras! PIP3 SH2 p85 p110 SH2 Recruitment from cytoplasm to PM, via SH2 domains SH2 p85 p110 How do we know proximity is not enough? 1. p85 mutants that activate without binding to RTKs 2. Tethering to membrane does not activate PI3-K pathway and Cancer Syndromes RTK GF Cancer Syndromes p PIP3 PI3-K PTEN Akt1/2 (Tuberous Sclerosis Complex) (Target of rapamycin) S6K Lipid PTPase Ser/Thr Kinase Hamartin Tuberin TSC2 TSC1 (Ras-homology enriched in brain) Lipid Kinase Ras GAP RheB Small GTPase mTOR Kinase 4EBP-1 Protein synthesis Cell growth/size/survival Kinase GI, Brain, Ovarian Cowden’s, Multiple Pancreas TSC Inhibitor of eIF4E Kovich & Cohen (2004) Dematology Online Journal 10: 3. Perelman (2004) Dematology Online Journal 10: 17. PIP3 targets include many GEFs, many tyrosine kinases, and others, including . . . PKB (aka Akt) = ser/thr kinase that promotes cell survival P P P P PIP3 PKB (= membrane lipid) PH K . . . is inactive in cytoplasm . . . contains a PH (pleckstrin homology) domain & a kinase domain Multi-step activation of PKB: proximity PIP3 P P PH P K PH domain recognizes 3’phosphate of PIP3, bringing kinase domain to the PM P Proximity to PM alone does not activate the kinase PH K Multi-step activation of PKB: covalent modification PIP3 P P PH P P Inactive PKB PDK1* K P P PH P P K P P Active (phos’d) PKB *PDK1 is also recruited to the membrane via a PIP3-binding PH domain Overall, two proximity steps plus (at least) one phosphorylation step EGFR Activation of PLCg combines THREE inputs P . . P P P PIP3 P P P P PIP2 P P PLCg (Inactive, in cytoplasm) PH SH2 1. PROXIMITY: Recruitment from cytoplasm to PM, via SH2 domains SH2 Catalytic EGFR Activation of PLCg combines THREE inputs 3. PROXIMITY: Binds to PIP3 via PH domain P . . P P P P P P P PIP2 DAG P SH2 P 2. COVALENT: Activated by EGFR phosph’n PH SH2 P Catalytic InsP3 Summary: Many RTK effectors require two or more simultaneous inputs for activation PI3K: recruitment via SH2, allosteric regulation by EGFR,p85 PKB: recruitment, phos’n by non-EGFR-kinase(s) PLCg: recruitment, phos’n, retention at PM by binding PIP3 Why multiple inputs to each effector? RTKs activate a complex network of interacting response pathways (and this is the simple version!) Active P RTK P P P STAT PI3K PLCg DAG InsP3 Rac STAT~P PKC Targets CaMK PI3K SOS ROS PTP Ras Cdc42 MAPK JNK Targets Targets Targets Targets Targets Targets Targets Targets Targets Targets Nuclear Transcription Factors PDK1 S6K PKB GSK3 Apoptosis Growth Factors and Receptor Tyrosine Kinases • • • • • RTK’s--How do they work? EGFR signaling and ras MAP Kinase Cascades PI3K, PKB, PLCg PTPs (Protein Tyrosine Phosphatases) But how do you shut these things off? Family of Protein Phosphatases Tonks & Neel, Curr Op Cell Bio (2001) How Do PTPs dephosphorylate specific targets? Intracellular targeting: “zip code” model Extra domains on PTPs confer localization and protein-protein interactions Initially thought that catalytic domains possessed little specificity for RTKs. However, co-crystal structures and biochemistry reveal that some PTPs catalytic domains exhibit exquisite sensitivity PTP-1B critical residues interact with Insulin Receptor T-loop residues Salmeen, et al Mol Cell (2000) PTEN opposes PI3K by removing PI3-phosphate PTEN discovered as a tumor suppressor gene. Mutated in brain, breast and prostate cancers. Has homology to dual specificity phosphates, but shows little activity toward phosphoproteins. Was discovered to remove phosphates from PIPs; thereby providing likely mechanism for tumor suppression. Cantley & Neel, PNAS (1999) Gleevec--proof that you can target kinases for drug therapy Goldman & Melo, NEJM, Oct 9, 2003 Gleevec--proof that you can target kinases for drug therapy