PosterMay - University of South Alabama
advertisement
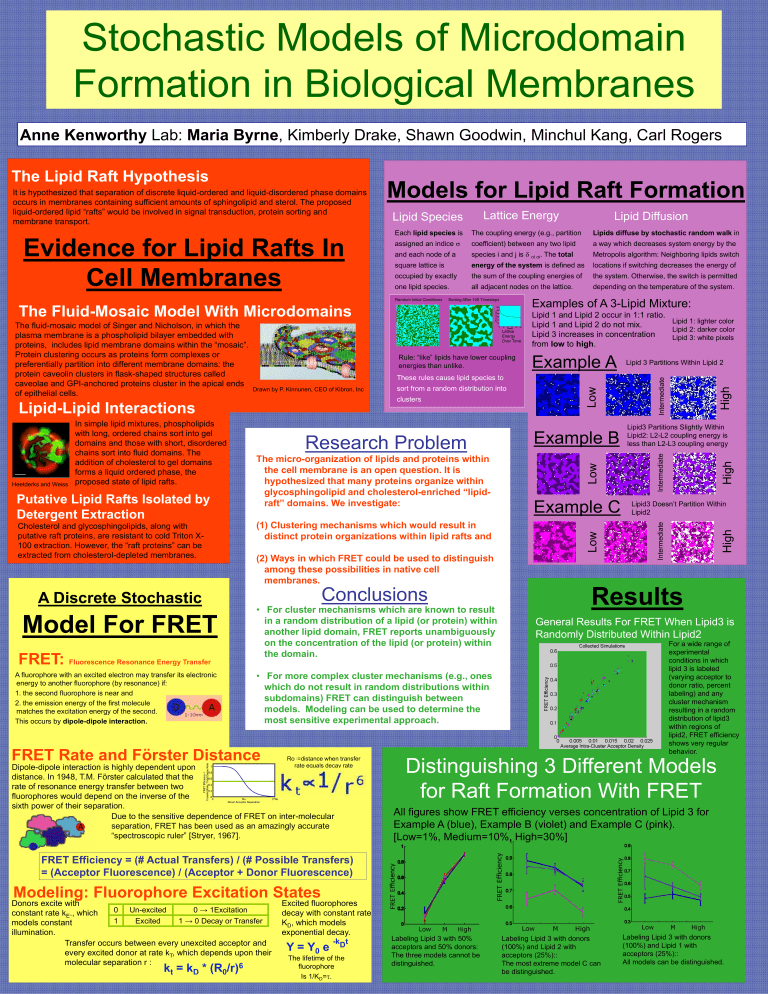
Stochastic Models of Microdomain Formation in Biological Membranes Anne Kenworthy Lab: Maria Byrne, Kimberly Drake, Shawn Goodwin, Minchul Kang, Carl Rogers Lattice Energy Lipid Species Each lipid species is The coupling energy (e.g., partition Lipids diffuse by stochastic random walk in assigned an indice coefficient) between any two lipid a way which decreases system energy by the and each node of a species i and j is i i. The total Metropolis algorithm: Neighboring lipids switch square lattice is energy of the system is defined as locations if switching decreases the energy of occupied by exactly the sum of the coupling energies of the system. Otherwise, the switch is permitted one lipid species. all adjacent nodes on the lattice. depending on the temperature of the system. Sorting After 100 Timesteps Examples of A 3-Lipid Mixture: Rule: “like” lipids have lower coupling energies than unlike. clusters Putative Lipid Rafts Isolated by Detergent Extraction Cholesterol and glycosphingolipids, along with putative raft proteins, are resistant to cold Triton X100 extraction. However, the “raft proteins” can be extracted from cholesterol-depleted membranes. A Discrete Stochastic Model For FRET FRET: Fluorescence Resonance Energy Transfer A fluorophore with an excited electron may transfer its electronic energy to another fluorophore (by resonance) if: 1. the second fluorophore is near and 2. the emission energy of the first molecule matches the excitation energy of the second. This occurs by dipole-dipole interaction. Research Problem The micro-organization of lipids and proteins within the cell membrane is an open question. It is hypothesized that many proteins organize within glycosphingolipid and cholesterol-enriched “lipidraft” domains. We investigate: (1) Clustering mechanisms which would result in distinct protein organizations within lipid rafts and Example B Low Heetderks and Weiss sort from a random distribution into Example C Low Drawn by P. Kinnunen, CEO of Kibron, Inc Low These rules cause lipid species to Lipid-Lipid Interactions In simple lipid mixtures, phospholipids with long, ordered chains sort into gel domains and those with short, disordered chains sort into fluid domains. The addition of cholesterol to gel domains forms a liquid ordered phase, the proposed state of lipid rafts. Example A (2) Ways in which FRET could be used to distinguish among these possibilities in native cell membranes. Conclusions • For cluster mechanisms which are known to result in a random distribution of a lipid (or protein) within another lipid domain, FRET reports unambiguously on the concentration of the lipid (or protein) within the domain. Ro =distance when transfer rate equals decay rate Dipole-dipole interaction is highly dependent upon distance. In 1948, T.M. Förster calculated that the rate of resonance energy transfer between two fluorophores would depend on the inverse of the sixth power of their separation. Due to the sensitive dependence of FRET on inter-molecular separation, FRET has been used as an amazingly accurate “spectroscopic ruler” [Stryer, 1967]. Lipid3 Partitions Slightly Within Lipid2: L2-L2 coupling energy is less than L2-L3 coupling energy Lipid3 Doesn’t Partition Within Lipid2 Results General Results For FRET When Lipid3 is Randomly Distributed Within Lipid2 For a wide range of experimental conditions in which lipid 3 is labeled (varying acceptor to donor ratio, percent labeling) and any cluster mechanism resulting in a random distribution of lipid3 within regions of lipid2, FRET efficiency shows very regular behavior. • For more complex cluster mechanisms (e.g., ones which do not result in random distributions within subdomains) FRET can distinguish between models. Modeling can be used to determine the most sensitive experimental approach. FRET Rate and Förster Distance Lipid 3 Partitions Within Lipid 2 High Lattice Energy Over Time Lipid 1 and Lipid 2 occur in 1:1 ratio. Lipid 1: lighter color Lipid 1 and Lipid 2 do not mix. Lipid 2: darker color Lipid 3 increases in concentration Lipid 3: white pixels from low to high. High The Fluid-Mosaic Model With Microdomains High Random Initial Conditions The fluid-mosaic model of Singer and Nicholson, in which the plasma membrane is a phospholipid bilayer embedded with proteins, includes lipid membrane domains within the “mosaic”. Protein clustering occurs as proteins form complexes or preferentially partition into different membrane domains: the protein caveolin clusters in flask-shaped structures called caveolae and GPI-anchored proteins cluster in the apical ends of epithelial cells. Lipid Diffusion Intermediate Evidence for Lipid Rafts In Cell Membranes Models for Lipid Raft Formation Intermediate It is hypothesized that separation of discrete liquid-ordered and liquid-disordered phase domains occurs in membranes containing sufficient amounts of sphingolipid and sterol. The proposed liquid-ordered lipid “rafts” would be involved in signal transduction, protein sorting and membrane transport. Intermediate The Lipid Raft Hypothesis Distinguishing 3 Different Models for Raft Formation With FRET All figures show FRET efficiency verses concentration of Lipid 3 for Example A (blue), Example B (violet) and Example C (pink). [Low=1%, Medium=10%, High=30%] FRET Efficiency = (# Actual Transfers) / (# Possible Transfers) = (Acceptor Fluorescence) / (Acceptor + Donor Fluorescence) Modeling: Fluorophore Excitation States Donors excite with 0 Un-excited 0 → 1Excitation constant rate kE., which 1 Excited 1 → 0 Decay or Transfer models constant illumination. Transfer occurs between every unexcited acceptor and every excited donor at rate kT, which depends upon their molecular separation r : kt = kD * (R0/r)6 Excited fluorophores decay with constant rate KD, which models exponential decay. -kDt Y = Y0 e The lifetime of the fluorophore Is 1/KD=. Labeling Lipid 3 with 50% acceptors and 50% donors: The three models cannot be distinguished. Labeling Lipid 3 with donors (100%) and Lipid 2 with acceptors (25%):: The most extreme model C can be distinguished. Labeling Lipid 3 with donors (100%) and Lipid 1 with acceptors (25%):: All models can be distinguished.