BMI.Core.ITHS.MEBI590.Fall.2007
advertisement

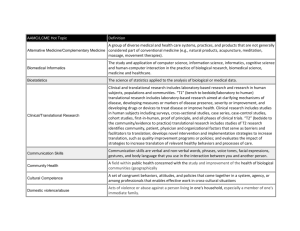
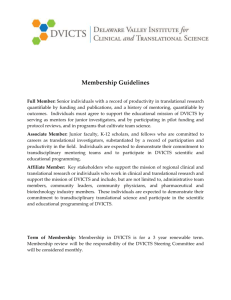
October 2, 2007 Biomedical and Health Informatics Lecture Series Peter Tarczy-Hornoch MD Head and Professor, Division of Biomedical and Health Informatics University of Washington Biomedical and Health Informatics Lecture Series Focus: current topics and developments in informatics Presenters: faculty, students, researchers and developers from UW, other academic institutions, government, and industry (locally and nationally) Intended audience: Broader UW & Seattle community interested in BHI BHI faculty and students History: Early 1990’s: initiated as part of IAIMS (MEDED 590) 2003-2006: temporarily changed to closed journal club format Fall 2006: return to public lecture series format Fall 2007: 10th year of Division of Biomedical & Health Informatics MEBI 590 & BHI Lecture Series Biomedical and Health Informatics (BHI) Lecture series available for credit as MEBI 590 Details & upcoming lectures available at: http://courses.washington.edu/mebi590/ pth@u.washington.edu Key points for those taking for credit Need to sign in each lecture to get credit CR/NC course Must attend 9 of 10 lectures for credit Informatics and the New Northwest Institute of Translational Health Sciences Peter Tarczy-Hornoch MD Director, Biomedical Informatics Core Northwest Institute of Translational Health Sciences Head and Professor, Division of Biomedical and Health Informatics Professor, Division of Neonatology bhi.washington.edu Outline Clinical Translational Science Awards Northwest Institute of Translational Health Sciences Biomedical Informatics Core of NW ITHS Data Integration Summary NIH Roadmap - Process Initiated in 2002 by NIH Director (Zerhouni) http://nihroadmap.nih.gov/ Chart a roadmap for medical research in 21st c. NIH Leadership What are today’s scientific challenges? What are the roadblocks to progress? What do we need to do to overcome roadblocks? What can’t be accomplished by any single Institute – but is the responsibility of NIH as a whole Working Groups Implementation Groups Implementation Groups => RFAs Summer/Fall 2006: New initiatives (Roadmap 1.5) NIH Roadmap – Themes New Pathways to Discovery Building Blocks, Biological Pathways, and Networks Molecular Libraries & Molecular Imaging Structural Biology Bioinformatics and Computational Biology (BISTI/NCBC) Nanomedicine Research Teams of the Future High-Risk Research Interdisciplinary Research Public-Private Partnerships Re-engineering the Clinical Research Enterprise Clinical Research Networks/NECTAR Clinical Research Policy Analysis and Coordination Clinical Research Workforce Training Dynamic Assessment of Patient-Reported Chronic Disease Outcomes Translational Research (Clinical Translational Science Awards) NIH Roadmap Clinical Translational Science Awards Initial request for applications October 2005 Current RFA: RFA-RM-07-007 CTSA planning grants (one year), implementation grants (five years) “The purpose of this initiative is to assist institutions to create a uniquely transformative, novel, and integrative academic home for Clinical and Translational Science that has the resources to train and advance a cadre of well-trained multiand inter-disciplinary investigators and research teams with access to innovative research tools and information technologies to promote the application of new knowledge and techniques to patient care.” Definition of Translational Research “Translational research transforms scientific discoveries arising from laboratory, clinical or population studies into clinical or population-based applications to improve health by reducing disease incidence, morbidity and mortality Modified from the NCI translational research working group (2006) UW: human subjects, specimens or plans CTSA: From Bench to Bedside to Community NIH Roadmap Clinical Translational Science Awards Integrate existing Clinical Research Centers (CRCs) with existing clinical/translational science training grants (K12, K30, T32) and expand capabilities through new cores (e.g. Biomedical Informatics, Evaluation, Novel Technologies, etc.) Establish regional and national consortia with the aim of transforming how clinical and translational research is conducted, and ultimately enabling researchers to provide new treatments more efficiently and quickly to patients When fully implemented in 2012, the initiative is expected to provide a total of about $500 million annually to 60 academic health centers in the US National CTSA Awards 2006 & 2007 CTSA Full Center Awards 2006 Columbia University Health Sciences Duke University Mayo Clinic College of Medicine Oregon Health & Science University Rockefeller University University of California, Davis University of California, San Francisco University of Pennsylvania University of Pittsburgh University of Rochester University of Texas Health Science Center at Houston Yale University 2007 Case Western Reserve University Emory University Johns Hopkins University of Chicago University of Iowa University of Michigan University of Texas Southwestern Medical Center University of Washington University of Wisconsin Vanderbilt University Washington University Weill Cornell Medical College Outline Clinical Translational Science Awards Northwest Institute of Translational Health Sciences Biomedical Informatics Core of NW ITHS Data Integration Summary Institute of Translational Health Sciences Northwest ITHS is the name for the regional inter-disciplinary consortium funded through the NIH-NCRR Clinical Translational Science Award (CTSA) Planning grant: 2006-7 Full Center grant: 2007-12 funded $62M NW ITHS will provide an “academic home” and integrated resources to: Advance clinical and translational science; Create and nurture a cadre of well-trained clinical investigators; Speed translation of discoveries into clinical practice Foster interactions between the university, non-profit, and business research communities Create an incubator for novel ideas and collaborations that cross disciplines Institute of Translational Health Sciences NW ITHS – “Collaboratory” Model NW ITHS - Partners Founding Members of the NW ITHS and Key Collaborators University of Washington Children’s Hospital and Regional Medical Center Fred Hutchinson Cancer Research Center Group Health Cooperative Center for Health Studies Benaroya Research Institute PATH Six proposed American Indian and Alaska Native Network Sites 6 Health Sciences School, 12 sites, 67 key scientific personnel, more than 150 centers Drs. Nora Disis (UW), Bonnie Ramsey (CHRMC), Mac Cheever (FHCRC/SCCA) co-leaders Institute of Translational Health Sciences Eleven ITHS Cores Administrative Novel clinical and translational methodologies Pilot and collaborative translational and clinical studies Biomedical informatics Study design and biostatistics Regulatory knowledge, support and research ethics Participant clinical interactions resources (CRC+) Community engagement Translational technologies and resources Research education, training and career development Tracking and evaluation Institute of Translational Health Sciences Outline Clinical Translational Science Awards Northwest Institute of Translational Health Sciences Biomedical Informatics Core of NW ITHS Data Integration Summary CTSA RFA & Biomedical Informatics Biomedical Informatics is the cornerstone of communication within (CTSAs) and with all collaborating organizations Applicants should describe: support provided for operations, administration, research and clinical/translational research activities plan to establish communication with external organizations relevant to their mission the process by which standards and other mechanisms will be developed and used to maximize interoperability between internal systems and systems in outside organizations assessment of informatics performance across the CTSA programs and with external partners inter- and intra-organizational sharing of data, technology and best practices Biomedical Informatics is expected to be the subject of an overall NIH CSTA Informatics Steering Committee that ensures interoperability between the CTSA institutions and with their external partners. Biomedical Informatics Core Team Peter Tarczy-Hornoch MD, Core Director Jim Brinkley MD PhD, Core Co-Director Nick Anderson PhD, Core Deputy Director Bill Lober MD Jim LoGerfo MD MPH Dan Suciu PhD Dan Ach (GCRC Informatics Lead) To be hired: ~14 professional staff and 3 RA slots ITHS Biomedical Informatics Core Aim 1 Aim 3 Aim 2 Aim 4 Aim 5: Develop & maintain ITHS administrative databases & Web interfaces Aim 1: Provide access to electronic health data at ITHS institutions Inventory and model recurring common queries Develop new interfaces to electronic health data from partner institutions Provide ITHS researchers access to electronic health data from partner institutions via a new common web interface Pilot a Virtual Data Warehouse (VDW) across the ITHS partner institutes building on the common web interface Extend the pilot VDW to include clinics in the WWAMI region Access to electronic health record data Existing resources: MIND Access Project (UW), Cerner Research Query System (CHRMC), Clinical Data Repository (FHCRC), Research-O-Matic (CHS) Gaps: no convenient access, repository data limited Goals: Simplify appropriate access to existing data Extend appropriate access to existing data Extend sources of electronic health record data Note: research still needed to solve Aim 1-4 gaps Aim 2: Support access to study data management tools for translational research Provide consultation to ITHS researchers regarding choosing and implementing study management tools Continue to develop and enhance existing ITHS data management tools Maintain and augment an inventory of data management tools Develop interfaces to most commonly use data management tools Perform a feasibility study of the establishment of a Data coordinating center Access to study data management tools Existing resources: GCRC Study Data Management (UW/CHRMC), Seedpod/Celo (UW), CF TDN (CHRMC), Clinical Informatics Shared Resource (FHCRC), multiple tools elsewhere Gaps: ease of use, limited features, not integrated Goals: Move local systems from prototype to production Develop centralized resources for currently used case report forms/study data management tools Extend centralized repository to include other CTSA tools Aim 3: Interface to biological study data from scientific instrumentation cores Provide ITHS researchers access to data from ITHS scientific instrumentation cores Prioritize list of other scientific instrumentation cores suitable to access Develop protocols and interfaces to new ITHS Human Genomics and Coordinated Tissue Bank core Access to instrumentation cores data Existing resources: large number of scientific instrumentation cores across consortium sites, generalizing interfaces via caBIG & SCHARP collaboration with Labkey Software (FHCRC) Gap: data not integrated with clinical/study data Goals: Build reusable interfaces to key scientific instrumentation Ensure compatibility with Aim 4 and national standards Aim 4: Integrate access across these three data sources Provide ad-hoc integration of aims 1-3 to ITHS researchers via ITHS BMI personnel Develop a data integration model for ITHS BMI by adapting existing tools Implement, test and refine prototype ITHS BMI Data Integration System Deploy and continue to refine the ITHS BMI data integration system Integrate access across these resources Existing resources: BioMediator (UW), XBrain (UW), CNICS, NA-ACCORD (UW), MIND/MAP (UW), Clinical Data Repository (FHCRC), caBIG (FHCRC), SCHARP (FHCRC), Virtual Data Warehouse (CHS) Gaps: no system integrates sources from Aim 1-3, no system across consortium members Goals: Adapt and evolve existing local systems to meet needs Continue to assess commercial systems Adopt interoperable approaches across CTSA sites Outline Clinical Translational Science Awards Northwest Institute of Translational Health Sciences Biomedical Informatics Core of NW ITHS Data Integration Summary UW Biomedical Data Integration and Analysis Research Group Peter Tarczy-Hornoch MD, PI Dan Suciu PhD, PI Alon Halevy PhD, Past PI 6 collaborating faculty Jim Brinkley, Chris Carlson, Eugene Kolker, Peter Myler, 4 programmers Ron Shaker, Todd Detwiler 13 students (over time) Eithon Cadag, Brent Louie, Terry Shen, Kelan Wang Motivation for Data Integration Genomics Data Literature Clinical Data Proteomics Information Pathways Knowledge Discovery (understanding) Experimental Data Others… Adapted from Chung and Wooley. 2003 Slide K. Wang, 2005 The Growth of Biologic Databases 900 800 700 Databases 600 500 400 300 200 100 0 2000 2001 2002 2003 2004 2005 2006 Year (Nucleic Acids Research, Database Issues 2000-2006) Slide E Cadag, 2006 BioMediator System Federated, general purpose, modular, decoupled NIH NHGRI/NLM funded 2000-2007 www.biomediator.org Interface Pfam Query` Query`` Query Translation Query Query` Interface Query`` ProSite Interface Query` Common data model CDD Query`` BioMediator Use Case: Annotation PubMed Entrez PROSITE COGs GO BLAST Human analysis and curation Local databases PSORT Pfam CDD BLOCKS Local algorithms Slide E Cadag, 2006 Finding Needle in Haystack: Inference Complete Result Set Relevant Subset Inference to Emulate Human Annotator Working memory Pfam.DomainHit IF DomainHit e-value > e-value: 10e-10 10e-15 name: neurotransmitter ProSite.DomainHit THEN remove e-value: 10e-20 name: neurotrans. IF DatabaseHit Name is BLAST.DatabaseHit similar to other e-value: 10e-10 DatabaseHit Names name: nic. acetylcholine THEN increase evidence BLAST.DatabaseHit e-value: 10e-20 evidence for name: acetylcholine acetylcholine increased rec. ... Rule-base ... Slide E. Cadag, 2006 Evaluation Scoring System Dimensions of granularity and utility Score Granularity Meaning Utility Meaning -2 Automated annotation is incorrect Phrasing or representation of automated annotation is not useful for functional annotation -1 Automated annotation is less specific than actual Automated annotation is less useful than actual 0 Automated annotation is indistinguishable from actual Automated annotation is as useful as actual Automated annotation is more specific than actual Automated annotation is more useful than actual +1 Slide E. Cadag, 2006 Scores for Automated Annotations Automated Score Incorrect or useless Less granular or useful Same as actual More granular or useful Total Granularity, % (n) Utility, % (n) 3.0% (1) 0% (0) 20.6% (7) 5.8% (2) 52.9% (18) 73.6% (25) 23.5% (8) 20.6% (7) 100% (34) 100% (34) Granularity average (selected annotations): -0.029 Utility average (selected annotations): 0.147 Slide E. Cadag, 2006 Finding Needle in Haystack: Uncertainty NSF IIS funded 2005-2009 Complete Result Set Relevant Subset Data Source Measures: Ps Source 1 Source 2 Concept 1 Concept 2 Source 3 Source 4 Concept 1 Concept 2 Ps: users belief in a concept from a particular source Slide B. Louie, 2007 Data Source Measures: Qs Source 2 Source 1 relationship Concept 1 Concept 2 Source 3 Source 4 Concept 1 Concept 2 relationship Qs: users belief in the interconnections (relationship) between two sources Slide B. Louie, 2007 Data Record Measures: Pr Source 1 Source 2 Concept 1 Concept 2 Record 1 Record 2 Pr: measure of belief in a particular data record Slide B. Louie, 2007 Data Record Measures: Qr Source 1 Source 2 Concept 1 Concept 2 link Record 1 Record 2 Qr: measure of belief in a particular link between data records Slide B. Louie, 2007 Result Graph with Uncertainty Measures Qs: 0.8 Qr: 0.9 Ps: 0.7 Pr: 0.3 Ps: 1.0 Pr: 0.8 Ps: 0.8 Pr: 0.5 Qs: 0.8 Qr: 0.3 Slide B. Louie, 2007 Network Reliability Theory Qse1* Qre1 UII (U2) Score = probability that a node is reachable from the start (seed) node. S Psn1* Prn1 Qse1* Qre1 Qse1* Qre1 Psn1* Prn1 Psn1* Prn1 Qse1* Qre1 Qse1* Qre1 Qse1* Qre1 Psn1* Prn1 Psn1* Prn1 Computing U2 score is #P. Approximation algorithms exist (Karger 2001), but are impractical. Qse1* Qre1 Slide B. Louie, 2007 Result Graph with Uncertainty Scores Qs: 0.8 Qr: 0.9 U2: 0.72 Ps: 0.7 Pr: 0.3 U2: 0.21 Ps: 1.0 Pr: 0.8 U2: 0.80 Ps: 0.8 Pr: 0.5 U2: 0.40 Qs: 0.8 Qr: 0.3 U2: 0.24 Slide B. Louie, 2007 BioMediator & Uncertainty: Evaluation Preliminary evaluation Gold standard: COG functional categorization Comparison: BioMediator + Uncertainty Agreement with actual: 94.4% After increasing number of simulations to estimate UII scores: 100% NW ITHS and Data Integration Aim 1 Aim 3 Aim 2 Aim 4 Aim 5: Develop & maintain ITHS administrative databases & Web interfaces Outline Clinical Translational Science Awards Northwest Institute of Translational Health Sciences Biomedical Informatics Core of NW ITHS Data Integration Summary Summary/Questions CTSAs are seen as a key part of the NIH Roadmap “Re-engineering the clinical research enterprise” Biomedical informatics (BMI) cores are seen as key nationally as well as locally for NW ITHS The BMI core is focused on addressing identified gaps through both research and tool development An important foundational element to the BMI core is data integration