“Facilitating Clinical and Translational Research Using Biomedical
advertisement

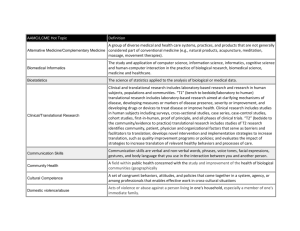
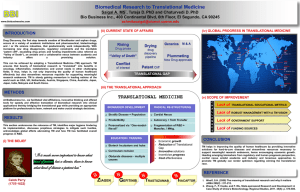
“Facilitating Clinical and Translational Research Using Biomedical Informatics” The vast majority of human diseases are the product of multi-step processes that involve the complex interplay of a multitude of genes acting at different levels of the genetic program. These genes are integrated and manifested as systemic phenotypes or medical findings detected during routine patient care or as part of clinical research studies. This fundamental observation has underscored the need to integrate basic science and clinical medicine more tightly, thus facilitating the rise of translational research as a distinct field in biomedicine. The goal of this field is to rapidly convert novel advances in basic science to improvements in patient care and to relay findings from clinical studies employing such new, and increasingly customized diagnostics and therapeutics back to the bench for further refinement of our understanding of the disease process. However, two significant roadblocks have been identified that make this translation difficult. The first translational roadblock (T1) is in the conversion of bench science findings to the development of tests for diagnosis, prognosis, or prevention or of customized therapies that may then be tested in humans. In contrast, the second translational block (T2) is in the transformation of results from clinical research studies to broadly accepted and implemented health care practices. While the former is focused more on the bench sciences and clinical research, the latter is focused on population-based studies. While addressing each of the translational blocks requires mastery of largely non-overlapping research disciplines (e.g. T1 requires the biological and clinical sciences, and T2 requires clinical epidemiology, behavioral science, public policy, communication theory, and qualitative research), the application and advancement of the inter-disciplinary field of biomedical informatics (BMI) is crucial for tackling both T1 and T2. BMI combines the quantitative disciplines of information and computer sciences and statistics with social sciences such as psychology and communication theory and the research and healthcare domains of bench science and clinical research, clinical medicine, and public health. Current translational blocks pertinent to informatics exist both within and between research, clinical, and government organizations such as academic institutions, intramural government research initiatives, inter-institutional clinical or research cooperatives, pharmaceutical companies, and healthcare providers. Namely, efficient knowledge transfer and access within and amongst these entities are severely hampered by the following: 1) Inability of informatics applications to syntactically interoperate (i.e. are ‘silos’ of data), forcing the end user to manage and transform data between tools. 2) Ineffective use of controlled vocabularies and standards-based, reusable common data elements that promote data integration and semantic interoperability of diverse population, clinical, biospecimen, imaging, and research data sets. 3) Lack of tools to facilitate data acquisition and storage using industry standard software design principles. 4) Unavailability of statistically sound software applications to co-analyze and visualize complex population and biomedical data sets (T1) or mathematically robust decision support tools (T2) using appropriate security measures and employing user-friendly interfaces that facilitate basic and advanced physician, clinician-scientist, or bench researcher workflows and processes. In this presentation, both local and nationwide/global solutions to address these roadblocks will be presented.