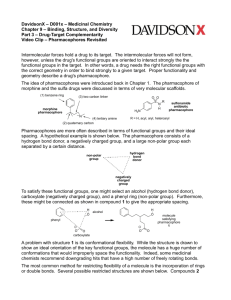
THROMBIN INHIBITOR WITH A RIGID TRIPEPTIDYL ALDEHYDES
advertisement

Formation continue chemoinformatique D. Hovarth , E. Kellenberger 2008/2009 PHARMACOPHORE TUTORIAL Aim: Introduction to pharmacophore generation and pharmacophore search We will first align three active molecules, create a pharmacophore based on common superimposed features and then we will perform virtual screening to identify potential hits in a chemical database Context: human Thrombin Thrombin (activated Factor II [IIa]) is a coagulation protein that has many effects in the coagulation cascade. It is a serine protease that converts soluble fibrinogen into insoluble strands of fibrin. It activates factor XI, factor V, and factor VIII and promotes platelet activation. Thrombin is produced by the enzymatic cleavage of two sites on prothrombin by activated Factor X (Xa). Prothrombin is produced in the liver and is post-translationally modified in a vitamin Kdependent reaction. Anticoagulants stop or delay blood from clotting and are medication for thrombotic disorders (thrombosis, pulmonary embolism, myocardial infarctions and strokes). Oral anticoagulants like warfarin act by antagonizing the effects of vitamin K, therefore impacting thrombin maturation. Direct thrombin inhibitors are a new class of anticoagulants. Some are already in clinical use, while others are undergoing clinical development. Several X-ray structures of the ligand-binding-domain of human thrombin in complex with inhibitors are available in the Protein Databank. Course material: All files are stored in the Pharmacophore directory. In INPUT subdirectory: - Thrombin inhibitors: active.mdb The database contains the active 3D structure of the co-crystal ligands extracted from 1c5n (1), 1ghv (2) and 1c1u (3) PDB entries. 1 - Databases: 2 3 database100.mdb 10 true inhibitors of human thrombin 90 decoys (DUD dataset) database100-conf.mdb 1-250 conformations /molecules in the database100.mdb In SCREEN subdirectory: - Superimposed thrombin inhibitors: ligand_ali.mdb The database contains the overlaid active 3D structure of compounds 1, 2 and 3. - Pharmacophore 5points.ph4 5 points pharmacophore build using the overlaid actives structure of compounds 1, 2 and 3. - Database100_ph.mdb hits selected in the database100.mdb upon pharmacophore search Software: EXERCISES MOE package, CCG To generate 3D coordinates in the database100-conf.mdb: MOE | File | Open database100.mdb MOE DBV | Compute | Energy Minimize In Database Minimize window, select Rebuild3D and switch on the Preserver Existing Chirality option. Then press OK to start calculation Don’t start calculation ! To generate a set a pool of 3D conformers as in database100-multiconfs.mdb: MOE | File | Open Import Conformations database100.mdb set up the conformer generation and the database output name (for instance database100-multiconfs.mdb). 1. 3D Alignment of thrombin inhibitors MOE | File | Open ligands.mdb In the MOE database viewer, select the 3 compounds (Ctrl and click left) and right click one of these and click Copy Selected to MOE in the popup menu. Close the Database Viewer. MOE | Compute | Conformations | Flexible Alignment In the Flexible Alignment window, lower the iteration limit to 20 and the Failure limit to 5. Do not change the other options. Start calculation by pressing OK. The calculation lasts for several minutes. A new database named flexalgn.mdb is created. It includes the possible alignments. Open ligands_ali.mdb database and compare the overlay of compounds 1, 2 and 3 in the different MOE-alignment solutions. Copy one alignment in MOE main GUI and close the all Database Viewer. 2. Pharmacophore query generation MOE | New | Pharmacophore Query In the Pharmacophore query editor window, give a title to the Query (e.g. thrombin). It is possible to create the query either manually or automatically. Choose the second option by clicking on Consensus.... In the Pharmacophore Consensus windows, select MOE as input, then click on calculate. A set of pharmacophore features is displayed into the MOE main GUI. Select three to five key features and click on Load Selected. Close the window. In the Pharmacophore query editor window, save your query as thrombin.ph4. Close the window. 3. Pharmacophore search MOE DBV | File | Phamacophore search In the Query section of the Pharmacophoric Search window, Browse… the thrombin.ph4 query. Click on Search to start the calculation. Report results in Table 1 (RIGID SEARCH). The search only retrieves few true hits, because only one conformer is considered for each molecule of the database. We will therefore generate a new database that contains a set of conformations for each compound. MOE | File | Open - In the Open window, select database100.mdb and click on Import Conformations. - In the Conformation Import window, set the number of Conformations in the Limits options to 25. Start calculation (It lasts for few minutes). - A more complete database of conformers, database100-multiconf.mdb, can be used instead. Repeat the pharmacophoric search, again using thrombin.ph4 pharmacophore query. MOE DBV | File | Phamacophore search Reports results in Table 1 (FLEXIBLE SEARCH). Modify the pharmacophore to increase the distance tolerance then repeat the database query. MOE DBV | File | Phamacophore search - In the Pharmacophore Search window, Click on Edit… near the thrombin Title. - In the Pharmacophore Query Editor window, click on each feature and increase the corresponding tolerance Å by changing R value. Try increase of 1Å - It is possible also to require that only partial match of the pharmacophore - Some pharmacophoric points must be edited because they are over complicated. For instance, some points can at the same time hydrophobes, aromatic, acceptor and donor! # of hits # of true hits # of false negative RIGID SEARCH Features: Tolerance: FLEXIBLE SEARCH Features: Tolerance: FLEXIBLE SEARCH Features: Tolerances: Table1: Pharmacophoric search results. Questions: - Using pharmacophore search, is it possible to discriminate the known thrombin inhibitors from decoys? Is the screening approach efficient? - What are, in the current tutorial, the main limitations of the pharmacophore search?