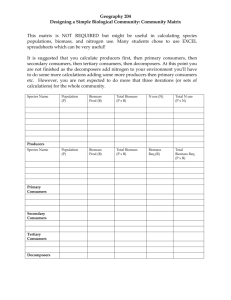
Plant allometry: is there a grand unifying theory
advertisement

871 Biol. Rev. (2004), 79, pp. 871–889. f Cambridge Philosophical Society DOI : 10.1017/S1464793104006499 Printed in the United Kingdom Plant allometry: is there a grand unifying theory? Karl J. Niklas* Department of Plant Biology, Cornell University, Ithaca, New York, 14853, USA (E-mail : kjn2@cornell.edu) (Received 10 June 2003 ; revised 16 April 2004; accepted 19 April 2004) ABSTRACT The study of size and its biological consequences – called allometry – has fascinated biologists for centuries. Recent advances in this area of study have stimulated a renewed interest in these scaling phenomena, especially in terms of the search for mechanistic explanations that transcend mere descriptive phenomenology. These advances are reviewed in the context of plant biology. Allometric derivations are presented that predict how annual growth in total body biomass is partitioned to construct new leaf, stem, and root tissues at the level of an individual plant. Derivations are also presented to predict annual reproductive effort and to predict how the biomass of body parts changes as a function of the number of plants per unit area in communities. The predictions emerging from these derivations are then examined empirically by comparing predicted and observed scaling exponents for each relationship using a world-wide data compendium gathered from the primary literature. These comparisons provide strong statistical support for each of the allometric predictions. This support is taken as evidence that a general unifying allometric theory for plant biology is near at hand. Nevertheless, the validation of this theory requires much additional work and raises a number of procedural and conceptual concerns that must be resolved before a single ‘ global ’ theory is accepted. Key words : annual plant growth rates, biomass allocation, body mass, community structure, density effects, organ partitioning, plant reproductive effort, size-correlated changes. CONTENTS I. II III. IV. V. VI. VII. VIII. IX. X. Introduction ................................................................................................................................................. The allometric formula : uses and abuses ................................................................................................. Derivations for organ biomass partitioning ............................................................................................. Derivations for annual growth rates of vegetative organs ..................................................................... Derivations for reproductive biomass ....................................................................................................... Derivations for plant community properties ........................................................................................... Assessing organ biomass partitioning and annual growth ..................................................................... Assessing reproductive effort ...................................................................................................................... Assessing plant community relations ........................................................................................................ Is there a single unifying theory? .............................................................................................................. (1) Size-dependent biases and ad hoc hypotheses .................................................................................... (2) Outliers: do exceptions prove a rule ? ................................................................................................ (3) Proportionality ‘ constants’ are not constant ..................................................................................... (4) Scaling exponents : fractions or numbers ? ........................................................................................ (5) Different ‘first principles ’ can yield the same predictions ............................................................... XI. Conclusions .................................................................................................................................................. 872 873 874 877 877 878 878 881 883 884 885 886 886 887 887 887 * Address for correspondence : Karl J. Niklas, Department of Plant Biology, Cornell University, Ithaca, New York, 14853, USA. Tel. : +01 607 255 8727 ; fax : +01 607 255 5407. Karl J. Niklas 872 XII. List of symbols .............................................................................................................................................. XIII. Acknowledgements ...................................................................................................................................... XIV. References .................................................................................................................................................... I. INTRODUCTION Size is one of the most important features of any organism. Well before the time of Darwin, natural historians observed and recorded manifold changes in plant and animal shape attending ontogenetic growth in size at the level of the individual. By the 1920s, these changes were mathematically explored by Julian S. Huxley (1924, 1932) who showed that the relative growth rates of different body parts can be deduced from the slopes of log–log plots of organ versus total body size. During the decades that followed, physiologists explored how metabolic rates varied as a function of body size across taxa from mice to elephants. The expectation was that metabolic rate would remain proportional to the 2/3 power of body size, reflecting the simple scaling relationship between body surface areas with respect to body volumes. However, the work of Samuel Brody and Max Kleiber revealed surprisingly that the slope of the log–log plots of metabolic rate versus body size was closer to 3/4 rather than 2/3 (Brody, 1945; Kleiber, 1947). The emergent 3/4 scaling ‘rule ’ was subsequently found to govern many other phenomena,includingsomerelatingtoplants(Hemmingsen,1960). Since the seminal works of Huxley, Kleiber, and Hemmingsen, the study of size and its consequences – called allometry – has burgeoned in scope and approach. Numerous physiological, morphogenetic, ecological, and evolutionary size-correlated trends for plants and animals have been observed, archived, and discussed conceptually (see McMahon & Bonner, 1983 ; Peters, 1983 ; Calder, 1984; Schmidt-Nielson, 1984; Niklas, 1994b ; Brown & West, 2000). Many of these trends appear to hold true across organisms as phyletically and ecologically diverse as microbes and trees or mosses and whales. These broad interspecific size-correlated trends are undeniably useful. Regardless of the mechanisms underlying them, they permit the description (and thus potential prediction) of many important biological relationships, at least within the boundary conditions proscribed by their statistical properties. Accordingly, the study of allometry is justifiable on the grounds of strict empirical enquiry. However, the importance of the study of allometry extends beyond description or prediction. If certain trends are size-dependent and ‘ invariant ’ with regard to phyletic affinity or habitat, they draw sharp attention to the existence of properties that are deeply rooted in all, or at least most living things. Identifying these properties using a first principles approach, therefore, has become something of a Holy Grail in biological allometry because any successful theory would unify as many diverse phenomena in biology as Einstein’s general theory of relativity has for physics. It is understandable, therefore, that numerous attempts have been made to provide an all-inclusive, unifying theory for broad interspecific allometric trends. However, most have 888 888 888 not held up against well-reasoned criticism or withstood empirical tests. For example, McMahon (1973, 1980) argued that metabolic rates increase as the 3/4 power of body mass because the cross-sectional areas of structural members must scale as the 3/4 power of the weight they support. Unfortunately, this explanation has little or no bearing on unicellular organisms or aquatic life forms for which the 3/4 power ‘ rule ’ also appears to hold true. Alternatively, Gray (1981) proposed that local variations in body temperature affect metabolism such that metabolic rates remain proportional to the 3/4 power of body mass. Yet, it is hard to imagine that ‘ local ’ variations in the body temperature of bacteria and unicellular algae are substantial. Equally provocative, Blum (1977) suggested that metabolic rates depend on the functional surface areas of organisms, which vary in time as well as in three dimensions. Noting that the surface area of a hypervolume increases as a function of volume raised to the 1xnx1 (where n is the number of dimensions), the 3/4 ‘ rule ’ immediately follows mathematically provided that n=4. Unfortunately, Blum (1977) never fully explored the ramifications of this idea (nor did he note that the variance for the numerical values of the slopes of metabolism versus body mass includes 0.80 such that organisms may exist in a five-dimensional space-time continuum). However, no attempt at a unifying allometric theory is more comprehensive or far-reaching than the theory proposed by Geoffrey West, James Brown, and Brian Enquist (1997, 1999, 2000), which purports to explain everything from Kleiber’s mouse-to-elephant 3/4 ‘ law ’ to the packaging of species in communities. Although similar to Blum’s (1977) hypervolume explanation, this theory argues that allometric relationships are governed by 1/4 power rules (or multiples of 1/4) because all organisms have internalized fractal delivery networks for energy and mass transfer, which have evolved to minimize the energy and time required to absorb, distribute and deliver resources internally. Although criticized on empirical and theoretical grounds and challenged by alternative conceptual approaches (see Banavar, Maritan & Rinaldo, 1999; Dodds, Rothman & Wertz, 2001 ; Darveau et al., 2002 ; Weibel, 2002), the WestBrown-Enquist theory currently remains the most comprehensive, so much so that it may be called a ‘ theory for everything. ’ The goal and scope of this article are far more circumspect. Here, my objective is to explore plant-size-correlated trends that are explicable in terms of two empirically undeniable trends. The first of these is that, across vastly different species and habitats, annual growth in biomass at the level of the individual plant scales as the 3/4 power of body mass. The second is that annual growth remains proportional to the capacity of the individual to harvest sunlight as judged by either cell pigment content or leaf biomass (Niklas & Plant allometry Enquist, 2001). These two size-correlated trends in conjunction with a comparatively small number of assumptions, each of which is predicated on simple biophysical principles, are used to predict the scaling exponents for four important aspects of plant allometry: (a) the partitioning of total body biomass among leaves, stems, and roots (Enquist & Niklas, 2002), (b) the annual growth rates for new leaf, stem, and root tissues (Niklas & Enquist, 2002 a, b), (c) the biomass annually invested in reproductive effort at the level of individual plants (Niklas & Enquist, 2003), and (d) a number of ecologically important properties of mixed or monotypic plant communities (Enquist & Niklas, 2001 ; Niklas, Midgley & Enquist, 2003 a ; Niklas, Midgley & Enquist, 2003b). The predictions of these derivations are then tested by examining a large database for plant organ biomass relations gathered from the primary literature. This foray into plant allometry is preceded by a brief treatment of the statistical protocols used in allometric analyses. This treatment is necessary because the interpretation of any size-dependent relationship relies on regression analysis to obtain the proportional (scaling) relationship between biological variables (which is given by the numerical value of the slope of the regression curve) and the proportionality factor governing the relationship (which is given by the Y-intercept of the regression curve). Thus, every empirical study of allometry and every theory for allometric relationships rests ultimately on the edifice of statistical inference. As we shall see, this edifice is remarkably robust and sophisticated, but it is nevertheless not as firm as one would like, largely because alternative regression models exist that can often obtain different slopes and constants depending on the nature of the data at hand (or the conceptual biases of the researcher). II. THE ALLOMETRIC FORMULA : USES AND ABUSES The term ‘ allometry’ was first coined by Julian S. Huxley (1924, 1932) who evaluated a large number of size-correlated trends and proposed that each could be approximated by the power function Y=bXa, where Y denotes the size of a body part (as gauged by its length or mass), X is a comparable measure of the size of the organism minus the size of the body part of interest, b is the allometric constant, and a is the allometric (scaling) exponent. Huxley’s formula is typically expressed logarithmically as log Y=log b+a log X (Gould, 1966 ; Smith, 1980 ; Sokal & Rohlf, 1981 ; LaBarbera, 1986, 1989 ; Niklas, 1994a). In this form, we see that log b is the Y-intercept (the numerical value of log Y when log X=1) and a is the slope of log Y versus log X (Sokal & Rohlf, 1981). Provided that the values of Y and X are measured for an individual organism over its ontogeny, for any unit time interval t, the change in the logarithm of Y equals a times the change in the logarithm of X, i.e. d (log Y)/dt=ad (log X)/dt such that dY/dX=aY/X (Huxley, 1932). The scaling exponent a is thus the ratio of the relative growth rate in the size of Y with respect to the relative growth rate in the size of X. 873 Huxley (1932) presented this formula as a true biological ‘law ’ – one that provides an a priori ‘ theory ’ for understanding the effects of size on morphometry. However, several workers have argued for a more pluralistic approach to the study of relative size, one that does not rest exclusively on the power formula log Y=log b+a log X (e.g. Smith, 1980 ; Harvey, 1982; Chappell, 1989). Indeed, Huxley’s first detractor was the individual to whom he dedicated his 1942 book, D’Arcy Thompson. Reevaluating the data sets used by Huxley, Thompson (1942) quickly showed that simple linear equations often fit some of Huxley’s untransformed data sets as well as or better than Y=bXa power functions and that linearly correlated pair-wise data remain linear when log-transformed. Indeed, many authors fail to provide a sound theoretical basis for accepting the power function relationship, and reevaluations of their data indicate that statistical analysis of untransformed data can give equally good results as that of the transformed data (e.g. Smith, 1980). Another problem in allometric analyses is the predictability of log–log regression curves, especially in the case of interspecific comparisons, which obtain high coefficients of correlation suggesting that Y values can be accurately predicted based on their corresponding X values. This problem arises when the variation in Y is large, as is often the case. Under these circumstances, the predictive capacity of log–log regression curves is remarkably low and analyses of residuals and per cent prediction errors are required. Another problem with using log-transformed data is that regression techniques fit a line to the mean values of the Y variable, but the mean of log-transformed variables is the median of the log-normal distribution (Gould, 1966 ; Sokal & Rohlf, 1981). Therefore, without correction, values reported for the antilog of the Y-intercept of the regression line are consistently biased (Prothero, 1986; Niklas, 1994 b). The correction factor, however, is simple ; it can be estimated from the standard error of log b. Unfortunately, many workers fail to report this important statistical parameter. Nevertheless, there are at least three good reasons for using log-transformed data. First, transformation of many forms of data typically reduces the problem of working with outliers ; second, log-transformed data typically comply with the statistical assumptions of normality and homoscedacity (Kermack & Haldane, 1950 ; Sokal & Rohlf, 1981) ; and, third, it provides a convenient means of examining proportionality that is unaffected by the unit of measurement, since the slope of the log–log regression line (log y2xlog y1)/ (log x2xlog x1) becomes ( y2/y1)/(x2/x1) when converted to a linear scale. For these reasons, power functions taking the form Y=bXa have become the traditional ‘ theory ’ in contemporary allometric analyses (see Gould, 1966 ; Peters, 1983 ; Calder, 1984 ; Schmidt-Nielson, 1984). A far more controversial issue is the type of regression analysis used in allometric analyses, because the type of analysis can profoundly influence the numerical values of scaling exponents and thus the extent to which observed exponents accord statistically with those predicted by a particular allometric theory (Sokal & Rohlf, 1981; Seim, 1983 ; Niklas, 1994 b). Based on standard statistical Karl J. Niklas 874 Table 1. Estimates of the allometric constant b and scaling exponent a for the formula Y2=b+aY1, where Y and b are log-transformed Regression method OLS Expression to be minimized n X ð y2i xbxay1i Þ2 Estimate of b Estimate of a y2 x ay1 sy2 y1 sy21 y2 x ay1 1 2sy2 y1 i=1 MA n X ( y2 xbxay1 )2 i 1+a2 i=1 RMA i n X ( y2 xbxay1 )2 i i=1 i a y2 x ay1 sy2 sy1 sy 2 sy1 ( 1=2 ) 2 sy22 sy21 + sy22 sy21 +4sy2 y 2 1 if sy2 y1 > 0 if sy2 y1 < 0 sy21 , sample variance of Y1 ; sy22 , sample variance of Y2 ; sy22 y1 , sample covariance of Y2 and Y1 ; OLS, ordinary least-squares regression analysis ; MA, major axis regression analysis ; RMA, reduced major axis regression analysis ; y1 and y2, numerical values of interdependent variables Y1 and Y2, respectively. inference, ordinary least-squares (Model Type I) regression analysis (denoted here as OLS) can be used provided that (a) the error term (log e) is normally distributed with a mean of zero and constant variance, (b) the distribution of log Y is normal at each value of log X, (c) the variance of log Y is constant across the range of log X, and (d) log X is an independent variable whose values are known without error (Sokal & Rohlf, 1981). Unfortunately, these four assumptions rarely if ever hold true for many data sets subjected to allometric analyses. Certainly, there is no ‘ independent variable’ when the size of the body parts of the same organism are compared. For these reasons, many allometricists have turned to Model Type II regression analyses, e.g. major axis (MA) or reduced major axis (RMA) regression analyses (see Harvey & Mace, 1982 ; Rayner, 1985; LaBarbara, 1986 ; McArdle, 1988; Niklas, 1994b). Model Type II analyses typically identify the variables of interest as Y2 and Y1 rather than Y and X to indicate their biological and functional interdependence. The choice of which kind of Model Type II regression analysis to use nevertheless remains. Each Model Type II regression protocol involves different assumptions about the error structure and variance relations between Y2 and Y1 to estimate the allometric constant b and the scaling exponent a (Table 1). In terms of selecting which Model Type II protocol to use, it is instructive to compare the assumptions underlying MA or RMA. The former is sensitive to the absolute measurement scales for the variables of interest and it is not especially robust to rotating the coordinate axes, whereas RMA is insensitive to both of these concerns. RMA is also less sensitive to assumptions about the error structure in a data set and it is less biased in terms of estimating the functional (allometric) relationship between two dependent variables. For these reasons, RMA has become one of the standard regression techniques for allometric analyses, although arguments can be made for MA (see Harvey & Mace, 1982 ; Rayner, 1985 ; LaBarbara, 1986 ; McArdle, 1988; Niklas, 1994 b). Importantly, the numerical value of the scaling exponent for the RMA regression line (aRMA) can be computed for the exponent of the corresponding OLS regression line (aOLS) using the formula aRMA=aOLS/r, where r is the OLS correlation coefficient. Likewise, the numerical value of the Y-intercept of the RMA regression curve (log bRMA) can be computed using the formula log bRMA=log Ŷ 2xaRMA log Ŷ1, where Ŷ denotes the mean value of Y. Methods for calculating the 95% confidence intervals of aRMA and bRMA are available ( Jolicoeur & Mosimann, 1968 ; Sokal & Rohlf, 1981; Rayner, 1985; McArdle, 1988), although each tends to give a highly conservative estimate of the intervals and thus inclines a worker to commit Type I error (the rejection of a valid allometric hypothesis). Provided that the coefficient of correlation r2 equals or exceeds 0.95, the choice of regression model is largely moot, since OLS, MA, and RMA obtain nearly equivalent numerical values for a. However, when r2<0.95, either Model Type II regression protocol (i.e. MA or RMA) gives higher numerical values for a than OLS. Throughout this article, RMA regression analyses are used to compare predicted with observed allometric trends (see Sections VII–IX). These comparisons emphasize the numerical values of scaling exponents, i.e. the slopes of log–log linear RMA regression curves, because this parameter emerges directly from allometric derivations for plant-size-correlated trends (see Sections III–VI). By contrast, the derivations to be reviewed do not predict the numerical values of allometric constants, which can be taxon-specific and can vary as a function of habitat. III. DERIVATIONS FOR ORGAN BIOMASS PARTITIONING In this and the following two sections, I present mathematical derivations that predict the scaling exponents, denoted by a, for a number of size-correlated trends. Plant allometry 875 A 3 2 1 0 −1 Log G T −2 −3 −4 −5 −6 −7 −8 −9 −10 −11 −12 −13 −16 −15 −14 −13 −12 −11 −10 −9 −8 −7 −6 −5 −4 −3 −2 −1 0 1 2 3 4 2 3 Log M T B 3 2 1 0 −1 Log G T −2 −3 −4 −5 −6 −7 −8 z algal species −9 herbaceous species and juveniles of woody species −10 −11 −12 −13 −16 −15 −14 −13 −12 −11 −10 −9 x woody species −8 −7 −6 −5 −4 −3 −2 −1 0 1 Log H Fig. 1. Log–log bivariate plots of total annual growth rate in body mass GT versus total body mass MT(A) and the capacity to harvest sunlight H(B). Lines denote reduced major axis regression curves for entire data set (A) and for portions of the data set (B). Scaling exponents and allometric constants for these relationships provided in Table 2. Data gathered from the primary literature (see text). The corresponding allometric constants for each relationship are denoted by b. Since the derivations in each section are extensive, successive allometric constants are denoted by sequentially numerical subscripts, i.e. b1, b2, etc. All of these derivations are predicated on two empirically well-established allometric trends (Fig. 1). Specifically, across a broad spectrum of aquatic and terrestrial nonvascular and vascular plant species, annual growth in plant body biomass (net annual gain in dry mass per individual, GT) remains proportional to the 3/4 power of total body mass (total dry mass per individual, MT) and directly proportional (scales isometrically with respect) to the capacity of an individual plant to harvest sunlight H as measured by the concentration of photosynthetic pigments per algal cell or the standing leaf biomass of a vascular plant with woody, nonphotosynthetic stems (see Niklas & Enquist, 2001). The evidence for these two trends comes from an analysis of three data sets, one for algal species (Niklas, 1994 b), one for tree-sized species (Cannell, 1982), and another recently compiled data set for small nonwoody species and juvenile Karl J. Niklas 876 Table 2. Statistical comparisons between predicted and observed relations among annual total body growth rates GT, total body mass MT, light-harvesting capacity H and standing leaf biomass ML. Scaling exponents and allometric constants are based on reduced major axis regression (aRMA and bRMA) of log10-transformed data (original units for growth rates are kg dry mass plantx1 ; original units for H and ML are kg dry mass plantx1). In all cases, P<0.001 or less. ++, prediction accepted ; +, prediction accepted because scaling exponent reflects differences in bRMA among data sets ; x, reject prediction ; CI, confidence intervals aRMA¡S.E. Y1 versus Y2 Predicted Antilog bRMA¡S.E. Observed 95 % CI Observed r2 n Within the size range of the entire data base : 5.65r10x16 kg f MT f 3.18r103 kg GT versus MT 0.75 0.76¡0.004 0.75–0.77 0.59¡0.017 0.97 1173 GT versus H 1.00 0.82¡0.003 0.81–0.70 2.05¡0.020 0.99 453 x16 x10 Within the size range of the algal data base : 5.65r10 kg f MT f 1.88r10 kg GT versus MT 0.75 0.75¡0.007 0.73–0.76 0.12¡0.100 0.99 66 GT versus H 1.00 1.05¡0.045 0.95–1.14 637¡0.647 0.89 66 Within the size range of the entire vascular plant data base : 5.93r10x6 kg f MT f 3.18r103 kg GT versus MT 0.75 0.66¡0.006 0.65–0.68 0.38¡0.017 0.90 1107 GT versus ML 1.00 0.83¡0.008 0.81–0.85 2.05¡0.021 0.96 387 Within the size range of the Cannell (1982) vascular plant data base : 4.16r10x1 kg f MT f 3.18r103 kg GT versus MT 0.75 0.77¡0.030 0.71–0.84 0.26¡0.066 0.80 132 GT versus ML 1.00 1.03¡0.039 0.95–1.12 1.59¡0.036 0.76 175 Within the size range of the new vascular plant data set : 5.93r10x6 kg f MT f 6.41r10x2 kg GT versus MT 0.75 0.72¡0.011 0.69–0.75 0.39¡0.028 0.79 975 GT versus ML 1.00 0.98¡0.021 0.95–1.03 5.51¡0.070 0.90 212 trees that have not accumulated substantial quantities of secondary tissues in stems and roots, which has been gathered from the primary literature published between 1992 and 2002 (Table 2). When the three data sets are pooled together in various ways, the scaling exponents are observed to depart, often significantly, from either 3/4 or unity but only as a consequence of group-specific differences in the allometric constant. For example, annual growth is observed to scale as the 0.82 – power of lightharvesting capacity across the entire pooled data base, but the one-to-one (isometric) relationship holds true when each of the three components of the data base is examined separately (Table 2). Thus, across all species, GT=b1MT3/4=b2H, where the numerical values of b1 and b2 are group-specific and may vary as a function of habitat. This relationship provides a basis for predicting the allocation of total body mass to leaf, stem, and root biomass for vascular plants with a stereotypical body plan (i.e. those consisting of leaves, stems, and roots). For these organisms, total vegetative body mass equals the sum of standing leaf, stem, and root biomass (ML, MS, and MR, respectively) such that GT=b1MT3/4=b1(ML+MS+MR)3/4. Likewise, since light harvesting capacity is a function of standing leaf biomass, the relationship GT=b2 ML holds true across species. Thus, M L =(b 1 /b2 )(M L +M S +M R ) 3/4=b 3 (M L +M S + MR)3/4. This last relation can be used to derive the individual scaling relationships among leaf, stem, and root biomass, provided a few assumptions are made (Enquist & Niklas, 2002; Niklas & Enquist, 2002 a ; Niklas, 2003). This derivation begins by noting that standing stem and root biomass must scale as some function of organ bulk F Prediction 35 624 62 737 ++ + 12 186 496 + + 9829 9495 + + 533 545 + + 3684 1993 + + tissue density r, organ diameter D, and organ length L. Denoting these stem and root features by the subscripts S and R, respectively, standing stem and root biomass can be expressed as MS=b4rSDS2LS and MR=b5rRDR2LR, respectively. Provided that stem and root bulk tissue densities are relatively constant for any species (i.e. b4rS=b6 and b5rR=b7), such that MS=b6DS2LS and MR=b7DR2LR, we see that ML=b3[ML+b6DS2LS+b7DR2LR]3/4. A critical assumption is required at this point in the derivation. Standing leaf biomass is assumed to scale as average cross-sectional stem and root area, i.e. ML=b8DS2=b9DR2. This assumption is biologically and physically reasonable. The mass of water flowing through stems and roots must be conserved and it must depend on the number and average cross section of water-conducting cell types (i.e. tracheids and vessel members), which are dependent on stem cross section (Carlquist, 1975 ; Zimmermann, 1983 ; Niklas, 1994b), particularly sapwood in woody species (Bond-Lamberty, Wang & Gower, 2001). Provided that ML=b8DS2=b9DR2 holds for any particular species, it then follows that ML=b3[1+(b6/b8)LS+(b7/b9)LR]3/4 (ML)3/4=b34[1+(b6/b8)LS+(b7/b9)LR]3. This last scaling relationship can be simplified further if it is assumed that average root and stem lengths scale isometrically with respect to one another (i.e. LR=b10LS). This assumption is based on the observation that, across diverse vascular species, stem and root meristematic growth and extension in organ length (and diameter) is coordinated, presumably through hormone-mediated developmental processes (e.g. Genard et al., 2001). If this assumption is true, then it follows that ML=b34[(1/LS)+(b6/b8)+(b7b10/b9)]3LS3. Plant allometry Noting that, for any species characterized by indeterminate growth in body size, 1/LSp0 as LS increases, if also follows that MLyb34[(b6/b8)+(b7b10/b9)]3LS3=b11LS3. Collectively, the foregoing scaling relationships indicate that MS=(b6/b111/3b8)MLML1/3=b12ML4/3, that MR= (b7b10/b9b111/3)MLML1/3=b13ML4/3, and that MS=(b12/b13) MR. It is also seen that the standing biomass of the shoot (the sum of all leaves and stems per plant) scales with respect to standing root biomass as ML+MS=(b12/b13)MR+ (MR/b13)3/4 (Enquist & Niklas, 2002). The allometric (taxon-specific) constants for these relationships may vary widely between different groups of plants as a consequence of phyletic or local environmental differences. However, in terms of their scaling exponents, the foregoing derivations indicate that leaf biomass should scale as the 3/4 power of stem (or root) biomass such that stem and root biomass will scale isometrically with respect to one another, i.e. ML ! MS3/4, ML !MR3/4, and MS ! MR. IV. DERIVATIONS FOR ANNUAL GROWTH RATES OF VEGETATIVE ORGANS The relations empirically observed among total annual growth GT, body mass MT, and standing leaf biomass ML (Niklas & Enquist, 2001) also permit the derivation of the relationships among the annual growth rates of leaves, stems, and roots, i.e. organ biomass production per plant per growth season (denoted as GL, GS, and GR, respectively) (Niklas & Enquist, 2002 a, b). From first principles, total annual growth in vegetative body mass must equal the sum of the annual growth of leaves, stems, and roots such that GT=GL+GS+GR. As noted, across vascular plants with woody, nonphotosynthetic stems, GT=b2ML and, for deciduous species, it is reasonable to assume that annual leaf growth scales as an isometric function of standing leaf biomass (i.e. GL=b14ML) such that GT=(b2/b14)GL. It therefore follows that GL= [b14/(b2xb14)](GS+GR)=b15(GS+GR). However, for nondeciduous species, annual leaf growth is likely to scale as an isometric function of the difference between standing leaf biomass and the leaf biomass produced in the previous growth season (Ml), which in turn is may be assumed to be some function of ML. That is, GL=b16(ML x Ml) and Ml=b17ML, respectively. If these assumptions are correct, then we see that, for nondeciduous species, GL= b16(1 x b17)ML=b18ML and GT=(b2/b18)GL=b19GL, from which it follows that GL=(GS+GR)/(b19x1)=b8(GS+GR). Noting that GL=b15(GS+GR) and GL=b20(GS+GR) are mathematically (but not numerically) equivalent, the general expression for the relationship between leaf growth and stem plus root growth across all vascular plants is predicted to be GL=b15 or b20(GS+GR)=b21(GS+GR). This general formula can be used to derive the scaling relations among the growth rates of all three vegetative organs as follows. It is reasonable to assume that stem and root growth rates scale as some function of the average diameter and length of stems and roots, i.e. GS=b22rSDS2LS and GR= b23rRDR2LR, respectively (see Section III). This assumption 877 is predicated on the dependency of organ growth rates on standing organ biomass. It is also reasonable to assume that the average bulk tissue densities of stems and roots are relatively constant for any particular taxon, i.e. b22rS=b24 and b23rR=b25, such that GS=b24DS2LS and GR=b25DR2LR. Under these circumstances, GL=b21 (b24DS2LS+b25DR2LR). Assuming, once again, that average root cross-sectional areas scale isometrically with respect to average stem cross-sectional areas (i.e. DR2=b26DS2) and that root length scales isometrically with respect to stem length (i.e. LR=b27LS), we see that GL=b21(b24+b25b26b27) DS2LS=b 28DS2LS. Therefore, GL=(b 28/b24)GS=b129GS, GL=[b29/(b29xb21)]GR=b30GR, and GS=(b30/b29)GR= b131GR. Likewise, total shoot growth (leaf and stem growth per year) is predicted to scale with respect to root growth as GL+GS=b30GR+b31GR=b32GR. The numerical values of the allometric ‘ constants’ featured in these derivations are expected to vary among taxa and as a result of local environmental conditions. However, in terms of proportional relationships, annual leaf growth rate is predicted to scale isometrically with respect to stem or root annual growth rates, i.e. GL ! GS !GR, whereas the annual growth rate of the shoot in toto will scale isometrically with respect to the annual growth rate of roots, i.e. GL+GS ! GR. V. DERIVATIONS FOR REPRODUCTIVE BIOMASS As noted, for most seed plants, annual growth in body mass scales as the 3/4 power of total body mass, which equals the sum of standing leaf, stem, and root biomass. That is, GT=b33MT3/4=b33(ML+MS+MR)3/4, where b33 is a taxon-specific (allometric) constant. With relatively few assumptions, it is possible to derive a relationship between the annual biomass invested in reproduction MP and the partitioning pattern of vegetative organ biomass, provided that plants are large such that MP is not a substantial fraction of total body mass (Niklas & Enquist, 2003). This derivation begins by stressing that, with the exception of very small or annual monocarpic species, MP does not contribute significantly to MT, because reproductive body parts, even for many slow-growing conifer species, are typically shed in less than one year. By contrast, when a plant enters its reproductive phase of growth, GT is the sum of GL, GS, GR, and GP, where GP is annual reproductive growth, which requires an annual expenditure in metabolic production. Therefore, across large and perennial seed plant species, GL+GS+GR+GP=b33(ML+MS+MR)3/4. As noted, MS and MR each scale as the 4/3 power of ML, whereas GL, GS, and GR scale isometrically with respect to each other. In other words, MS=b34ML4/3, MR=b35ML4/3, GS=b36GL, and GR=b37GL (Enquist & Niklas, 2002 ; Niklas & Enquist, 2002 b). We also know that, on average, GL=b38 ML, where b6 includes units of yearx1 (Niklas & Enquist, 2002 a). Accordingly, GP=b33(ML+MS+MR)3/4x(1+b36+b37)b38 ML. Assuming that the relation between annual reproductive 878 growth and biomass scales isometrically as GP=b39MP, where b39 includes units of yearx1, the scaling relation between MP and ML, MS, or MR is MP=b40(ML+b41ML4/3)3/4 xb42ML, MP=b40[(MS/b34)3/4+(b41/b34)MS]3/4xb42(MS/ b34)3/4, and MP=b40[(MR/b35)3/4+(b41/b35)MR]3/4xb42 (MR/b35)3/4, where b40=b33/b39, b41=b34+b35, and b42= (1+b36+b37)(b38/b39). Each of these equations describes a slightly nonlinear log–log (concave) relation for MP versus ML, MS, or MR. However, as will be seen, each predicted trend is approximated reasonably well by a linear log–log relationship. Importantly, none of the scaling relations used to derive the scaling relationships between reproductive biomass and leaf, stem, or root biomass directly or indirectly relates MP to ML, MS, or MR. Therefore, no mathematical ‘circularity ’ exists if MP is predicted based on the values of ML, MS, or MR reported in the literature for plants (regardless of their reproductive status). The scaling exponent and the allometric constant for the relationship MP versus ML, MS, or MR depend exclusively on the numerical values of b40–42. In turn, these ‘ constants’ depend on the vegetative biomass partitioning pattern of a particular species or higher taxon. Although all of the foregoing derivations cannot predict the numerical values of the allometric constants a priori, they can be tested directly by evaluating whether predicted values for the scaling exponents and the allometric constants agreed with those observed for inter – and intraspecific reproductive trends (see Niklas & Enquist, 2003). VI. DERIVATIONS FOR PLANT COMMUNITY PROPERTIES Here, my focus shifts from the level of the individual to that of an entire plant community. Once again, the whole-plant annual growth rate GT is observed to scale as the 3/4 power of MT and isometrically with respect to the capacity to intercept sunlight H, i.e. GT=b43MT3/4 and GT=b44H1, where b43 and b44 are group-specific constants. For vascular plants, H is proportional to ML (Niklas & Enquist, 2001). Therefore, GT=b43MT3/4=b44H=b45ML and ML= b46MT3/4, where b46=b43/b45 (allometric constants, which may or may not vary across taxa). Based on hydraulic and other biophysical considerations, ML is assumed to scale isometrically with respect to stem cross-sectional area, which is proportional to the square of basal stem diameter DS2. Therefore, ML=b46 MT3/4=b47DS2. Assuming that the three-dimensional space ai occupied by an individual plant is proportional to basal stem crosssectional area DS2, i.e. ai=b48DS2, the maximum number of individual plants possible in a community per unit area sampled N (‘ plant density ’) equals the quotient of the total unit area occupied by all individuals AT and the area occupied by an average individual in any given community. Therefore, it mathematically follows that N=AT/ai= AT/b48DS2=(b47/b46 b48) AT/MT3/4=b49 AT/MT3/4 (Niklas et al., 2003 a). Because AT is a constant whenever comparisons across different communities employ equivalent sample areas, Karl J. Niklas i.e., AT equals some constant k, the preceding scaling relations take the form of N=k/b48DS2=(b47/b46b48)k/MT3/4 =b49k/MT3/4 such that, at the level of an individual plant, four proportional relations emerge, i.e. MT ! Nx4/3, GT ! Nx1, ML ! Nx1, and DS2 ! Nx1 (Enquist & Niklas, 2001; Niklas et al., 2003 a). As in all cases, these relationships are influenced by potential taxon-specific variation in biomass allocation, a variety of allometric constants, and by rates of limiting resource supply for a given environment. However, the scaling exponents for these relationships are expected to be insensitive to these factors. Finally, because the numerical value for each variable Y measured at the level of an individual plant equals the total community value for the variable Ŷ divided by N, i.e. Y=Ŷ/N, the scaling relationship for an entire community becomes Ŷ ! N1+a. Thus, from the foregoing proportional relationships, we see that total plant biomass M̂T is expected to scale as N 1x4/3 or N x1/3, whereas total community growth in biomass per year ĜT, total community standing leaf biomass M̂L, and total basal stem area D̂S2 are each predicted to scale as N 1x1 or N 0. Accordingly, at the level of entire plant communities, total standing biomass will scale as the x1/3 power of plant density, whereas total community growth, standing leaf biomass, and total basal stem area are expected to be invariant provided that communities have reached an equilibrium with their available resources (Niklas et al., 2003 a). VII. ASSESSING ORGAN BIOMASS PARTITIONING AND ANNUAL GROWTH To assess the predictions for standing leaf, stem, and root biomass and for annual leaf, stem, and root growth in biomass (see Sections III–IV), data for these parameters were gathered from the primary literature. For the majority of tree species, the bulk of these data comes from Cannell (1982) who compiled data sets for tree-sized dicot, monocot, and conifer species as well as a limited number of bamboo species (see Enquist & Niklas, 2002; Niklas and Enquist, 2002a, b). Each of the Cannell (1982) data sets is standardized to 1.0 ha and represents approximately 600 sites world-wide, published in a standardized tabular format that provides the primary citation and, when supplied by authors, longitude, elevation, the age of the dominant species (or conspecific in the case of monotypic managed stands), the number of plants per 1.0 ha (‘ plant density ’), height, total basal stem cross-sectional area, and the standing biomass and net biomass production of stem wood, bark, branches, fruits, foliage, and roots (in units of metric tons dry matter per year). The values for annual stem wood, bark, foliage, etc. production used here reflect as much as possible annual losses of dry matter due to mortality, litter-fall, decay, and consumption (see Cannell, 1982). Organ biomass and productivity were determined by authors from direct measurements of fully dissected representative plants (typically f5 individuals) for the majority of the Cannell (1982) sites. Authors regressed these data to Plant allometry 879 Table 3. Statistical comparisons between predicted and observed relations among standing leaf, stem, root, and shoot biomass per plant (ML, MS, MR, and MSH, respectively). Scaling exponents and allometric constants are based on reduced major axis regression (aRMA¡S.E. and bRMA¡S.E.) of log10-transformed data (original units for biomass are kg dry mass plantx1). In all cases, P<0.0001 or less. +, prediction accepted ; x, prediction rejected ; MT, total individual body mass ; CI, confidence intervals aRMA¡S.E. Y1 versus Y2 Predicted Antilog bRMA¡S.E. Observed 95 % CI Observed x6 r2 n Within the size range of the entire vascular plant data base : 5.93r10 kg f MT f 3.18r10 kg ML versus MS 0.75 0.75¡0.004 0.74–0.76 0.20¡0.011 0.97 1117 ML versus MR 0.75 0.84¡0.006 0.82–0.85 0.40¡0.015 0.96 678 1.00 1.12¡0.006 1.11–1.13 2.66¡0.012 0.98 736 MS versus MR MSH versus MR y1.00 1.07¡0.005 1.06–1.08 3.28¡0.014 0.97 1244 Within the size range of the Cannell (1982) vascular plant data base : 4.16r10x1 kg f MT f 3.18r103 kg ML versus MS 0.75 0.75¡0.008 0.73–0.76 0.12¡0.012 0.910 661 ML versus MR 0.75 0.79¡0.016 0.76–0.82 0.41¡0.016 0.861 338 MS versus MR 1.00 1.09¡0.009 1.05–1.13 2.59¡0.012 0.971 366 MSH versus MR y1.00 1.08¡0.014 1.05–1.11 3.54¡0.020 0.94 345 Within the size range of the new vascular plant data set : 5.93r10x6 kg f MT f 6.41r10x2 kg ML versus MS 0.75 0.87¡0.010 0.85–0.89 0.43¡0.035 0.94 456 ML versus MR 0.75 0.96¡0.016 0.92–0.99 0.74¡0.016 0.91 340 MS versus MR 1.00 1.03¡0.020 0.99–1.07 1.01¡0.066 0.86 370 MSH versus MR y1.00 1.07¡0.012 1.06–1.08 2.32¡0.041 0.89 899 estimate total organ biomass per 1.0 ha community sample. Data based on estimated regression variables were rejected when entering the Cannell (1982) data sets for the purposes of the analyses presented here and elsewhere (Enquist & Niklas, 2002; Niklas & Enquist, 2002a, b, 2003). Importantly, most of these data sets are for even-aged conspecific stands (n=600 out of 880 usable data sets) and biomass production values are typically averaged values for two or more years. Therefore, for each site used in the following analyses, the variance in standing organ biomass and biomass production was assumed to be comparatively small and annual production rates were considered representative of ‘ normal ’ rather than idiosyncratic growth seasons. Standing leaf, stem and root biomass per ‘average ’ plant was computed for each of the Cannell (1982) sites using the quotient of total community standing organ biomass and plant density. Annual leaf, stem, and root production rates were similarly calculated using the quotient of annual organ type production per community sample and plant density. However, it must be noted that most of the Cannell (1982) data sets probably underestimate standing root biomass and biomass production, particularly those of fine and small roots, because these are more difficult to excavate completely for increasingly larger root systems and because these fine and small roots are reported to increase disproportionately with increasing plant size (see Niklas & Enquist, 2002 a). Thus, numerically higher scaling exponents than those predicted were anticipated for any regression analysis using root biomass or biomass production as the variable plotted on the abscissa. Since the Cannell (1982) data sets emphasize mature and large plant body sizes, additional data were gathered from the primary literature published between 1990 and 2002 for species with comparatively small mature body sizes F Prediction 33 893 16 444 29 568 49 165 + x x + 8425 2439 13 621 5786 + + x + 7441 3371 2181 7297 x x x + 3 (e.g. species of Arabidopsis, Bromus, Lactuca, Lycopersicum, Plantago, Spartina), or for seedlings and saplings of tree species (e.g., Betula, Quercus, and Thuja) (see Table 2 for size range). These additional data, which come from 57 species not represented in the Cannell (1982) data sets, are from laboratory or field studies of plants grown under normal field or experimental conditions (e.g. elevated CO2, UV-B radiation, salinity, or soil micronutrient levels). The only criterion used to select data was that the variance per treatment was small as gauged by the standard errors reported for standing biomass or biomass production (Enquist & Niklas, 2002 ; Niklas & Enquist, 2002a, b ; Niklas, 2003). For these additional species, standing organ biomass and annual organ biomass production were calculated as for the Cannell (1982) data sets. Table 3 provides comparisons among the predicted and observed scaling exponents for the relationships among standing leaf, stem, root, and shoot (leaf and stem) biomass, i.e. ML, MS, MR, and MSH, respectively. The 95 % confidence intervals for the scaling exponent aRMA (the slope of the log–log linear reduced major axis regression curve) for each relationship provide an assessment of whether an observed scaling exponent complies statistically with that predicted. Provided that the 95% confidence intervals of the observed exponent include the predicted numerical value of the scaling exponent, the relevant allometric derivation is judged to be valid. In turn, the 95 % confidence intervals for the allometric constant bRMA for each relationship are used to assess whether there are statistically significant differences in the Y-intercepts of the log–log linear reduced major axis regression curves for different portions of the entire data set. If the 95 % confidence intervals of the allometric constant for a particular scaling relationship differ among the different portions of the entire data set, then comparisons of the Karl J. Niklas 880 A 2 1 0 log ML −1 −2 −3 −4 herbaceous species and juveniles of woody species −5 −6 −7 x woody species −6 −5 −4 −3 −2 −1 0 1 2 3 4 log MS B 2 1 log ML 0 −1 −2 −3 −4 −5 −6 −7 −6 −5 −4 −3 −2 −1 0 1 2 3 2 3 log MR log MS C D log MSH slopes of the regression curves for the different portions of the entire data set are used to assess the validity of the relevant allometric derivations. Inspection of Table 3 indicates that, across the entire data set, the scaling relations predicted for leaf biomass with respect to stem biomass and for shoot biomass with respect to root biomass agree with those predicted by the allometric derivations, whereas the scaling relations between leaf or stem biomass with respect to root biomass are not in agreement. This discrepancy appears to be the result of a statistically significant difference in the allometric constants of the regression curves for the Cannell (1982) data sets (predominantly tree-sized individuals) and the data from smaller sized plants (herbaceous species and juveniles of tree species) (Fig. 2). Regression analyses across the plant size range represented in the Cannell (1982) data sets (i.e., 4.16r10x1 kg f MT f 3.18r103 kg) reveals that the allometric derivations and observation are compatible in terms of the relationship between standing leaf and root biomass, whereas the relation predicted for standing stem versus root biomass remains incompatible with the trend in the data. Likewise, for smaller sized plants (i.e. 5.93r10x6 kg f MT f 6.41r10x2 kg), the predicted and observed scaling exponents for the relationships among standing leaf, stem and root biomass are incompatible. Nevertheless, isometric scaling exponents are observed for the relationship between shoot and root biomass (Table 3). The aforementioned discrepancies are reconciled in part by turning attention to the relations among annual organ growth in biomass, because standing organ biomass and annual organ growth in biomass are equivalent for very small herbaceous plants and for juveniles of tree-sized species that have yet to accumulate or produce secondary tissues (wood and periderm). During the first year of growth, standing stem biomass and annual stem growth are the same quantities, i.e. MS=GS. Likewise, standing root biomass and annual root growth in biomass are equivalent quantities, i.e. MR=GR. In successive years, however, secondary tissues (specifically, wood) accumulate in the stems and roots of woody species (see Franco & Kelly, 1998). Thus, over time, the biomass of standing stems and roots increases with age such that MS>GS and MR>GR. This feature of plant growth is relevant to the preceding comparisons of standing organ biomass, because the scaling relations for ML, MS, and MR are nearly isometric for the data drawn from small herbaceous plants and for juveniles in the size range of 5.93r10x6 kg f MT f 6.41r10x2 kg (Table 3). Indeed, the predictions of the allometric derivations for standing organ biomass emerge as statistically strong when they are placed in the context of annual leaf, stem, and root growth, i.e. GL, GS, and GR, respectively (Table 4). Specifically, comparisons of annual organ growth rates indicate that isometric scaling relations exist across the entire data set and within each of the two portions of the data set (tree-sized plants, and herbaceous plants and juveniles of tree-sized species). The single exception is the relationship between annual leaf and stem growth for tree-sized plants (i.e. 4.16r10x1 kg f MT f 3.18r103 kg), which may be the result of sampling biases or neglecting the effects of herbivory 4 3 2 1 0 −1 −2 −3 −4 −5 −6 −7 −7 −6 −5 −4 −3 4 3 2 1 0 −1 −2 −3 −4 −5 −6 −7 −7 −6 −5 −4 −3 −2 −1 0 1 −2 −1 0 1 log MR log MR 2 3 Fig. 2. Log–log bivariate plots for the relationships among standing leaf, stem, root, and shoot biomass (ML, MS, MR, and MSH, respectively). Lines denote reduced major axis regression curves. Scaling exponents and allometric constants for these relationships provided in Table 3. Data gathered from the primary literature (see text). (which would reduce standing leaf biomass). Since standing organ biomass and annual organ growth rates are equivalent for herbaceous and juvenile tree individuals, the absence of the 3/4 power relations predicted for leaf biomass with respect to stem or root biomass for small herbaceous species and juveniles of tree-sized species (see Table 3) is Plant allometry 881 Table 4. Statistical comparisons between predicted and observed relations among annual leaf, stem, and root growth rates (GL, GS, and GR, respectively). Scaling exponents and allometric constants are based on reduced major axis regression (aRMA¡S.E. and bRMA¡S.E.) of log10-transformed data (original units for growth rates are kg dry wgt./plt./yr.). In all cases, P<0.0001 or less. ++, prediction accepted ; +, prediction accepted because scaling exponent reflects differences in bRMA among data sets ; x, prediction rejected ; CI, confidence intervals aRMA¡S.E. Y1 versus Y2 Predicted Antilog bRMA¡S.E. Observed 95 % CI r2 Observed n Within the size range of the entire vascular plant data base : 5.93r10x6 kg f MT f 3.18r103 kg GL versus GS 1.00 0.98¡0.006 0.97–1.01 0.62¡0.012 0.97 709 GL versus GR 1.00 1.05¡0.010 1.03–1.06 1.82¡0.022 0.96 422 GS versus GR 1.00 1.17¡0.012 1.15–1.20 3.09¡0.025 0.96 402 x1 3 Within the size range of the Cannell (1982) vascular plant data base : 4.16r10 kg f MT f 3.18r10 kg GL versus GS 1.00 0.93¡0.015 0.90–0.96 0.62¡0.013 0.88 431 GL versus GR 1.00 1.00¡0.034 0.92–1.07 2.02¡0.023 0.77 205 Gi versus GR 1.00 1.02¡0.019 1.00–1.07 3.69¡0.013 0.93 205 Within the size range of the new vascular plant data set : 5.93r10x6 kg f MT f 6.41r10x2 kg GL versus GS 1.00 0.98¡0.018 0.94–1.01 0.71¡0.051 0.91 278 GL versus GR 1.00 1.02¡0.022 0.97–1.06 1.17¡0.065 0.90 217 GS versus GR 1.00 1.08¡0.031 1.00–1.13 0.88¡0.094 0.84 197 Analyses of the Cannell (1982) world-wide data base for seed plant biomass relations supplemented with data collected from the primary literature (see Section VII) provides robust statistical support for the allometric derivations for plant reproductive effort (Table 5). Across all species, the allometric constants emerging from these derivations (see Section V) are b40=0.027, b41=4.22, and b42=0.018. Inserting these values into the derivations for reproductive biomass indicates that MP should scale as the 0.861 power of standing leaf biomass ML and have an allometric constant (Y intercept ; bRMA) of 0.067 (Fig. 4A). This scaling relationship is statistically indistinguishable from that observed : aRMA= 0.841 and bRMA=0.064 (Table 5). Likewise, the scaling relations predicted for MP based on observed values of MS or MR were statistically indistinguishable from those observed statistically (Table 5 ; Fig. 4 B, C). In passing, the accuracy of the derivations for reproductive effort is comparable to that of direct regression analysis of the raw (non-transformed) data. The smallest difference between predicted and observed MP is observed when ML is used as the predictive variable for both methods of estimating reproductive biomass. Likewise, both methods underestimate MP for some of the largest tree species, perhaps because published values of MP for these species were measured by some authors after leaf-fall or herbivory. Nonetheless, the derivations are strikingly accurate for the majority of the plants in the database, even within the size range of small annual species (Fig. 4). log GL B log GL VIII. ASSESSING REPRODUCTIVE EFFORT A C log GS entirely consistent with the allometric derivations, especially in light of the differences in the numerical values of the allometric constants governing these relations (Table 4; Fig. 3). 2 1 0 −1 −2 −3 −4 −5 −6 −6 −5 −4 −3 2 1 0 −1 −2 −3 −4 −5 −6 −6 −5 −4 −3 2 1 0 −1 −2 −3 −4 −5 −6 −6 −5 −4 −3 F Prediction 21 559 10 184 9983 ++ + + 3238 671 2771 x ++ ++ 2692 1974 1024 ++ ++ ++ herbaceous species and juveniles of woody species x woody species −2 −1 0 1 2 −2 −1 0 1 2 −2 −1 0 1 2 log GS log GR log GR Fig. 3. Log–log bivariate plots for the relationships among annual growth rates of leaf, stem, and root biomass (GL, GS, and GR, respectively). Lines denote reduced major axis regression curves. Scaling exponents and allometric constants for these relationships provided in Table 4. Data gathered from the primary literature (see text). Karl J. Niklas 882 Table 5. Representative statistical comparisons between predicted and observed scaling exponents (aRMA) and taxon-specific (allometric) constants (bRMA) for inter- and intraspecific relations of reproductive, leaf, stem, and root biomass (MP, ML, MS, MR, respectively) based on reduced major axis regression of log10-transformed data (original units in kg of dry mass plantx1). In all cases, P < 0.0001. F- and r-values for predicted relations o85 000 and o0.998, respectively. CI, 95 % confidence intervals. See Section V for definitions of b40–42 aRMA¡S.E. 95 % CI Antilog bRMA¡S.E. Across all species (b40=0.027, b41=4.22, b42=0.018) MP versus ML Predicted 0.861¡0.002 0.856–0.865 0.067¡0.004 Observed 0.841¡0.025 0.784–0.898 0.064¡0.046 MP versus MS Predicted 0.657¡0.001 0.655–0.659 0.059¡0.003 Observed 0.674¡0.016 0.637–0.709 0.051¡0.039 MP versus MR Predicted 0.654¡0.001 0.652–0.656 0.049¡0.003 Observed 0.700¡0.020 0.656–0.745 0.044¡0.046 MP versus ML Across angiosperms (b40=0.023, b41=7.77, b42=0.015) Predicted 0.918¡0.003 0.912–0.923 0.101¡0.006 Observed 0.924¡0.035 0.858–0.990 0.115¡0.065 MP versus ML Across conifers (b40=0.036, b41=9.87, b42=0.056) Predicted 0.961¡0.001 0.958–0.963 0.161¡0.001 Observed 0.778¡0.072 0.515–1.042 0.167¡0.066 MP versus MS Within species Pinus rigida (conifer) (b40=0.142, b41=25.6, b42=0.001) Predicted 0.909¡0.002 0.905–0.909 1.66¡0.002 Observed 0.909¡0.015 0.876–0.942 1.66¡0.021 Dolicus lablab (dicot) (b40=0.090, b41=10.2, b42=0.002) Predicted 1.24¡0.002 1.23–1.24 21.9¡0.002 Observed 1.25¡0.081 1.06–1.42 22.1¡0.291 Pennisetum glaucum (monocot) (b40=0.024, b41=5.61, b42=0.017) Predicted 0.775¡0.002 0.774–0.776 0.235¡0.002 Observed 0.776¡0.017 0.739–0.810 0.232¡0.008 Significant numerical differences in b40 –42 are nevertheless evident between angiosperms and conifers as well as among individual species (Niklas & Enquist, 2003). These differences account for most of the ‘data scatter ’ observed in bivariant plots because, in each case, allometric derivations accurately predict all observed inter- and intraspecific MP trends (Table 5). For example, across all angiosperms, statistical analyses reveal that b40=0.023, b41=7.77, and b42=0.015 such that MP is expected to scale as the 0.918-power of ML with bRMA=0.101. These values are statistically indistinguishable from those observed. Similarly accurate results are obtained when the data from conifer species are examined (Table 5). The reproductive trends of phyletically and ecologically disparate species for which b40–42 values can be determined are also accurately predicted by the allometric derivations presented here (Fig. 5). For example, in the case of Pinus rigida, MP is predicted to scale as the 0.909 power of MS with bRMA=1.66, whereas aRMA=0.909¡0.015 and bRMA= 1.66¡0.021 are observed (Table 5). Likewise, for the large annual monocot species Pennisetum glaucum, MP is predicted to scale as the 0.775 power of MS with bRMA=0.235, whereas aRMA=0.776¡0.017 and bRMA=0.232¡0.008 are observed. n r2 F 0.068–0.069 0.057–0.072 279 279 — 0.754 — 851.1 0.048–0.049 0.047–0.055 418 418 — 0.754 — 1331 0.048–0.050 0.040–0.048 204 204 — 0.827 — 967.0 0.098–0.104 0.092–0.143 195 195 — 0.799 — 768.2 0.159–0.160 0.167–0.259 84 84 — 0.296 — 34.5 1.58–21.60 1.49–1.84 16 16 — 0.822 — 193.7 44 44 — 0.822 — 193.7 50 50 — 0.976 — 1920 95 % CI 21.7–22.1 5.71–85.5 0.233–0.237 0.227–0.237 These analyses suggest that tradeoffs are involved when a finite amount of total body mass is partitioned between or among different organ types (Enquist & Niklas, 2002 ; Niklas & Enquist, 2003). Clearly, the expenditures required to construct new leaf, stem, or root tissues annually (as gauged by their annual growth in biomass or by the difference in standing biomass between successive years) are removed from a pool of resources available for the construction of reproductive organs. That these tradeoffs have been reconciled differently by different species is evident from the numerical differences observed among the allometric constants b40–42 for different plant lineages (angiosperms versus conifers) or different individual species within each lineage of seed plant. These differences indicate that taxa with dissimilar biomass partitioning patterns with respect to their vegetative organs will have different allometric trends in their reproductive effort, whereas those with the same or very similar vegetative partitioning patterns will share similar b40 –42 values and thus share similar MP scaling relations. An important caveat is that reproductive effort can and has been measured by different authors using very different ‘ currencies, ’ e.g. seed number or biomass, and flower or fruit biomass. Likewise, it is not immediately obvious whether some portions of the plant body should be included when Plant allometry log MP B 2 1 0 −1 −2 −3 −4 −5 −6 −7 −6 A 1 x −5 −4 −3 −2 log ML −1 0 1 log MP 2 1 0 −1 −2 −3 −4 −5 −6 −7 −5 −1 −2 −3 −4 angiosperms conifers −5 −6 −6 2 B −5 −4 −3 −2 log M L −1 0 1 2 0 −1 −2 −5 −4 −3 −2 −1 0 1 2 3 4 P. glaucum P. rigida −3 −4 log MS C 2 0 log MP 2 1 0 −1 −2 −3 −4 −5 −6 −7 −6 log MP log MP A 883 −5 −5 D. lablab −4 −3 −2 −1 0 1 log M S −4 −3 −2 −1 0 1 2 3 log MR Fig. 4. Log–log bivariate plots for the relationships among reproductive biomass and standing leaf, stem, and root biomass (MP, ML, MS, and MR, respectively) for angiosperm and conifer species. Solid lines denote reduced major axis regression curves of data ; dashed lines denote predicted relationships based on allometric derivations for predicting MP. Scaling exponents and allometric constants for predicted and observed relationships are provided in Table 5. Data gathered from the primary literature (see text). estimating reproductive effort, e.g. should the scale-bract complexes of conifer megasporangiate cones be assigned a ‘ vegetative ’ or ‘ reproductive ’ organ status; are the pedicles of flowers ‘vegetative ’ or ‘ reproductive ’ organs? The data used here to measure reproductive effort undoubtedly reflect a broad range of ‘currencies ’ depending on the protocols used by different authors (see Körner, 1994, for a discussion regarding vegetative biomass fractionation protocols). An additional concern is that none of the data examined here incorporate the biomass of pollen grains, which are clearly metabolically costly (Niklas, 1994 b). Therefore, although the derivations pertaining to reproductive effort appear to be remarkably robust in terms of predicting observed trends at the level of individual plants, Fig. 5. Log–log bivariate plots for the relationships among reproductive biomass and standing leaf and stem biomass (MP, ML, and MS, respectively) for angiosperm species (A) and three individual species (Dolicus lablab, a legume vine ; Pinus rigida, a conifer tree ; and Pennisetum glaucum, a herbaceous monocot) (B). Solid lines denote reduced major axis regression curves of data ; dashed lines denote predicted relationships based on allometric derivations for predicting MP. Scaling exponents and allometric constants for predicted and observed relationships are provided in Table 5. Data gathered from the primary literature (see text). the concordance between predicted and observed scaling exponents must be viewed cautiously and with some degree of skepticism. IX. ASSESSING PLANT COMMUNITY RELATIONS Statistical analysis of the broad database discussed in Section V provides strong support for the scaling relationships predicted for the biomass partitioning and annual growth rates of plants at the individual level and for the relationships between these parameters and the number of plants per unit area sampled (‘ plant density ’) (Table 6). As predicted, total plant biomass MT scales as the x1.27 power of plant density N (Fig. 6 A) ; the 95 % confidence intervals including x4/3 and excluding x3/2 (Table 6). Also meeting predictions, annual plant growth rate GT and standing plant leaf biomass ML scale isometrically with respect to N (a=x0.98 and x1.00, respectively), whereas basal stem diameter DS scales as the x0.53 power of N (Fig. 6B–D). Karl J. Niklas 884 Table 6. Predicted and observed scaling exponents aRMA and allometric constants bRMA for log10-transformed data from the Cannell (1982) compendium. In each case, P < 0.001 or less. ML, MR, MSH, standing leaf, root, and shoot biomass ; MT, total plant biomass ; GT, annual total body growth in biomass ; DS, basal stem diameter ; N, plant density Observed Log Y versus log X Predicted aRMA¡S.E. Across all communities Log MT versus log N x4/3 x1.27¡0.03 Log GT versus log N x1.0 x0.98¡0.03 Log ML versus log N x1.0 x1.00¡0.03 Log DS versus log N x1/2 x0.53¡0.01 Log MSH versus log N x4/3 x1.31¡0.03 Log MR versus log N x4/3 x1.17¡0.03 Across angiosperm-dominated communities Log MT versus log N x4/3 x1.30¡0.05 Log GT versus log N x1.0 x1.00¡0.05 Log ML versus log N x1.0 x1.00¡0.03 x1/2 x0.51¡0.02 Log DS versus log N Log MSH versus log N x4/3 x1.25¡0.04 Log MR versus log N x4/3 x1.08¡0.05 Across conifers-dominated communities Log MT versus log N x4/3 x1.34¡0.04 Log GT versus log N x1.0 x1.04¡0.03 Log ML versus log N x1.0 x1.12¡0.04 x1/2 x1.28¡0.05 Log DS versus log N Log MSH versus log N x4/3 x1.31¡0.03 Log MR versus log N x4/3 x1.39¡0.04 95 % CI log10 bRMA¡S.E. r2 n F x1.33 to x1.16 x1.03 to x0.92 x1.05 to x0.95 x0.56 to x0.51 x1.36 to x1.25 x1.23 to x1.10 5.96¡0.11 3.99¡0.09 3.78¡0.09 0.86¡0.04 5.99¡0.10 4.97¡0.11 0.801 0.861 0.669 0.673 0.753 0.773 342 205 670 792 668 347 1368 1257 1348 1624 2029 1172 x1.36 to x1.23 x1.11 to x0.90 x1.05 to x0.95 x0.55 to x0.48 x1.33 to x1.17 x1.18 to x0.98 5.74¡0.16 3.76¡0.15 3.37¡0.10 0.75¡0.06 5.81¡0.14 4.72¡0.16 0.759 0.833 0.751 0.685 0.733 0.729 174 74 331 342 325 178 542.2 359.9 993.1 740.1 888.3 465.9 x1.42 to x1.25 x1.11 to x0.98 x1.19 to x1.05 x1.37 to x1.19 x1.36 to x1.25 x1.46 to x1.31 5.96¡0.11 3.99¡0.09 4.41¡0.10 5.33¡0.16 5.99¡0.10 6.26¡0.13 0.846 0.880 0.746 0.827 0.753 0.782 168 131 339 169 668 343 909.3 949.8 992.1 800.8 2029 1220 The predicted scaling relationship for shoot biomass MSH versus N at the level of an individual plant is also supported. Specifically, MSH scales as the x1.31 power of N with 95% confidence intervals that include x4/3 and exclude x3/2 (i.e., x1.36 to x1.25) (Table 6). However, MR scales as the x1.17 power of N and in this case the 95% confidence preclude the predicted x4/3 value (Table 6). This discrepancy can be attributed to a systematic, size-dependent underestimation of fine and small root biomass, i.e. progressively larger plants have disproportionately more fine and small root biomass, which are systematically more difficult to excavate with increasing plant size (see Niklas et al., 2002). This bias is expected to elevate the numerical value of the scaling exponent for MR versus N. This conjecture is supported by the exponent for conifer MR versus N. Conifer MR, which tends to be more shallowly buried and thus more easily excavated than angiosperm MR, scales as the x1.28 power of N with 95% CI that include the predicted value of x4/3 (Table 6). The derivations presented for plant density relationships are also supported based on empirically observed total community mass-growth-density relationships (Table 7). For example, individual MT and GT are predicted to scale as the x4/3 and the x1 power of N, respectively, but total community biomass M̂T is predicted to scale as the as the x1/3 power of N, whereas total community growth ĜT is predicted to be independent of N. Regression of M̂T versus N and ĜT versus N gives scaling exponents of x0.266¡0.03 and 0.007¡0.03, which are statistically compatible with the predictions of the model (Table 7). Total community leaf biomass M̂L is also predicted to be independent of N, and the best fit regression curve for the observed data of M̂L versus N gives a scaling exponent statistically indistinguishable from zero. Similar comparisons show that the predicted and observed scaling exponents for other total community biomass-plant density relationships are statistically indistinguishable, although it is evident that the coefficients of correlation are low (Table 7). X. IS THERE A SINGLE UNIFYING THEORY ? This article outlines a set of previously published allometric derivations interrelating individual plant growth, vegetative and reproductive organ biomass partitioning, and a variety of density-correlated community properties. All of these derivations are predicated on two statistically robust observed trends, each of which is shown to span many orders of magnitude and a broad spectrum of taxonomically and ecologically divergent species. These trends are the 3/4 scaling relationship between total annual plant growth in biomass and total body biomass and the one-to-one (isometric) relationship between the capacity to harvest sunlight and body biomass (see Niklas & Enquist, 2001). The predictions emerging from these derivations are shown here and elsewhere to receive strong statistical support, as judged by comparing the numerical values of predicted scaling exponents and those observed for an extensive database accumulated from the published literature. However, this concordance must be viewed with a Plant allometry 885 log MT A 4 log GT B 3 2 1 0 −1 −2 −3 3 2 1 0 −1 −2 −3 −4 angiosperms conifers 2 3 2 3 4 log N 4 5 6 7 5 6 7 5 6 7 5 6 7 log N C 3 log ML 2 1 0 −1 −2 −3 log DS D 2 3 4 2 3 4 log N 0 −1 −2 log N Fig. 6. Log–log bivariate plots for the relationships among total body mass MT, total annual growth in biomass GT, standing leaf biomass ML, basal stem diameter DS, and plant density N (number of plants per unit sampled area) for mixedspecies and monospecific angiosperm and conifer-dominated communities. Lines denote reduced major axis regression curves of data. Scaling exponents and allometric constants for predicted and observed relationships are provided in Table 6 (see Table 7 for entire community relationships). Data taken from Cannell (1982) (see text). critical eye. Indeed, the study of allometry is intrinsically limited (if not outright defined) by the availability and the statistical structure of data just as it is rife with methodological and philosophical pitfalls. Space does not permit a systematic in-depth treatment of each of the problems facing the allometricist. However, some problems require special attention. (1 ) Size-dependent biases and ad hoc hypotheses Size-dependent biases are manifold within and across biological data sets. That these biases can have a profound effect on suppositions regarding allometric trends in terms of statistical techniques is obvious. What is less obvious is the temptation to erect ad hoc hypotheses in light of a theoretical framework to account for biases that may not actually exist. For example, the scaling exponent for the relationship between standing leaf and root biomass is predicted to be 3/4, whereas the lower 95% confidence interval for the observed scaling exponent exceeds 0.75 (see Table 3, Fig. 2). This discrepancy between observation and theory is explicable provided that standing root biomass is systematically underestimated with increasing body size. Larger root systems are arguably increasingly more difficult to excavate, whereas the biomass of small and fine roots is reported to increase disproportionately as total root biomass increases (see Makkonen & Helmissar, 2001). Likewise, feeder roots, which increase in number as a function of overall root system size, typically decompose annually (e.g. Niklas et al., 2002). Importantly, systematic size-dependent biases such as these will progressively ‘deflate’ reported values of standing root biomass for larger plants and thus artificially ‘inflate ’ the numerical values of the scaling exponents of bivariate regression curves whenever root biomass is plotted along the abscissa. However, it cannot escape attention that an alternative explanation is that the observed relationship between leaf and root biomass is in fact accurate. Indeed, no concrete evidence has been provided demonstrating that standing root biomass is systematically underestimated. Regardless of how ‘reasonable ’ an ad hoc hypothesis may appear, its plausibility does not equate with proof of existence. Unfortunately, some workers are so beguiled by a particular theory that they neglect the possibility that it may be wrong. A related concern is the ease with which mathematical post hoc hypotheses can be erected and the difficulties of testing them empirically. In this context, consider the many biophysically plausible assumptions that were required to simplify the otherwise intractable mathematical expressions for certain morphometric relationships. These assumptions give the appearance of being robust theoretically but they are nevertheless untested. For example, when treating the scaling relationships among standing leaf, stem, and root biomass, it was assumed that average stem diameter (and thus cross-sectional area) scales isometrically with respect to average root diameter (and cross section). The biophysical basis for this assumption is that the volume of water flowing per unit time from roots to stems must be conserved. Although the amount of water flowing from roots and through stems must be equivalent at the level of an individual plant, differences in the volume fractions and average cross-sectional areas of waterconducting cells in roots and stems undeniably exist across species. Thus, for some species, average root and stem diameters may not be equivalent for all species at the level of representative individuals (see Zimmermann, 1983; Gartner, 1995) especially for succulents and arid-adapted species (Holbrook, 1995 ; Niklas et al., 2002). Likewise, all of the derivations presented here assume that foliar leaves are the principal or sole light-harvesting organs, whereas it is clear that many species rely on photosynthetic stem tissues for capturing radiant energy (see Niklas, 2002; Pfanz et al., 2002). Taxon-specific differences in body plan Karl J. Niklas 886 Table 7. Predicted scaling exponents at the levels of the individual plant (see Table 6) and entire communities and those observed for total community mass–growth–density relationships Predicted Observed Individual Community a¡S.E. 95 % CI r2 n F MT ! Nx4/3 GT ! Nx1.0 ML ! Nx1.0 DS ! Nx1/2 MSH ! Nx4/3 MR ! Nx4/3 M̂T ! N x1/3 ĜT ! N 0 M̂L ! N 0 D̂S ! N 1/2 M̂SH ! N x1/3 M̂R ! N x1/3 x0.266¡0.03 0.007¡0.03 0.000¡0.03 0.467¡0.01 x0.310¡0.03 x0.165¡0.03 x0.33 to x0.16 x1.03 to 0.92 x0.05 to 0.05 0.42 to 0.51 x0.36 to x0.25 x0.23 to x0.10 0.135 0.000 0.000 0.610 0.143 0.034 342 495 675 792 668 347 53.28*** 0.002 0.000 1237*** 111.8*** 23.44** *** Significant at the 0.001 level. ** Significant at the 0.01 level. ML, MR, MSH, standing leaf, root, and shoot biomass ; MT, total plant biomass ; GT, annual total body growth in biomass ; DS, basal stem diameter ; N, plant density. architecture or physiological features thus clearly exist and may account for numerical differences observed in allometric constants. (2 ) Outliers : do exceptions prove a rule ? Here we turn to the question of statistical ‘outliers ’ in allometric trends. Many exist that are not immediately accounted for by the obvious differences in phylogenetic affinity that translate into body plan differences. This raises a number of related issues. One is the relative frequency of outliers, is it high or low with respect to species that appear to comply with a posited interspecific trend ? The analyses presented here indicate that comparatively few outliers exist for the majority of allometric trends explored. Nevertheless, if broad interspecific allometric trends are indicative of deeply rooted and fundamental biological phenomena, then data that deviate substantially from otherwise well-confined trends point to circumstances where general ‘ rules ’ are obviated in one manner or another. Clearly, outliers may be the result of unusual environmental circumstances or unusual taxon-specific features. In this regard, optimization theory indicates that organisms are likely to reflect form-function compromises within the context of their particular environmental circumstances (see Niklas, 1994 a). Every organism must perform manifold tasks to grow, survive, and reproduce. Many of these tasks have contradictory ‘best solutions’ and it is the reconciliation of these tasks that often leads to evolutionary diversification and the biodiversity we see today. For example, vertically oriented stems minimize the mechanical bending moments induced by self-loading, but they also minimize the amount of sunlight that can be captured daily. In a desert environment, columnar unbranched stems may be adaptive, whereas in other habitats the reverse may be true. The trilogy of form-function-environment is often neglected as is the fact that seemingly very different morphologies may be equally effective at performing all their functional obligations simultaneously in the same habitat. In this sense, allometric outliers are extremely instructive, even if they give the theorist momentary grief. Under any circumstances, if a ‘global ’ theory for size-dependent trends is advocated, understanding why ‘ outliers ’ exist (and what they tell us about biology) can only foster a deeper understanding of biodiversity in light of theory. ( 3) Proportionality ‘ constants ’ are not constant Yet another important concern is that allometric theory and practice are currently incapable of predicting a priori the numerical values of the allometric constants governing size-dependent trends. That these ‘ constants’ can numerically differ significantly for the same size-correlated relationship depending on the taxonomic group studied is selfevident from the analyses discussed here. The historical emphasis on scaling exponents in allometric theory and practice is understandable, since these exponents shed light on the proportional changes among correlated variables such as leaf and stem biomass or annual growth rates. But, from a biological perspective, the manner in which the absolute quantities of these variables change as a function of body size is equally important. Consider the hypothetical case of two plant species A and B for which leaf biomass ML scales as the 3/4 power of stem biomass MS but for which the allometric constants are b=1.5 and b=10, i.e. ML=1.5MS3/4 and ML=10MS3/4, respectively. Both of these hypothetical species evince the same proportional relationship, since a=3/4. However, for any value of MS, species B will have an order of magnitude more standing leaf biomass than species A. Indeed, differences in the values of allometric ‘constants ’ often account for much of the ‘data scatter ’ observed for broad interspecific allometric trends, because many of these trends are ‘composites ’ of numerous parallel intraspecific allometries sharing the same proportional relationships differing in their Y intercepts. Such differences in the numerical values of allometric constants probably reflect fundamental phylogenetic differences in how the representatives of different lineages are architecturally constructed. This aspect of allometric study awaits detailed empirical exploration and conceptual explanation. Plant allometry (4 ) Scaling exponents : fractions or numbers ? A serious concern is the inclination of allometricists to convert the numerical values of scaling exponents into fractions as they explore a favoured theoretical framework. Consider a hypothetical database that obtains a log–log regression curve with a slope of 0.79 and 95 % confidence intervals of 0.74 and 0.84. Is this slope indicative of a 3/4 scaling relationship, or a 4/5 or 5/6 relationship ? A strictly empirical approach to allometry can rely on statistical inference employing the numerical value of 0.79 within the boundary conditions of 0.74 and 0.84. However, the temptation to seek a mechanistic explanation may lead a particular worker to claim 0.79 as evidence for a 3/4, 4/5, or 5/ 6 ‘ rule ’ depending on the theoretical bias. Which, if any among these contending ‘ rules ’ is real must remain problematic unless the numerical values (and posited fractions) of the slopes of many different theoretically interrelated allometric trends can be used to eliminate some alternatives (e.g. Niklas, 2003). Yet, even this approach is not immune to the foibles of ‘ fractions versus numbers.’ For example, suppose that a certain allometric theory predicts that variable Y1 scales as the a1 power of variable Y2 and that variable Y2 scales as a2 power of variable Y3, i.e. Y1 ! Y2 a1 and Y2 !Y3a2 . It then follows that this theory makes three predictions that can be subjected collectively (rather than individually) to empirical enquiry, i.e. Y1 ! Y2 a1 , Y2 ! Y3 a2 , and Y1 ! Y3a1 a2 . But let us cast this hypothetical case in terms of regression analyses involving numbers rather than fractions. Suppose our theory predicts that a1=3/4 and a2=2 such that a1a2 is predicted to equal 3/2. Suppose in turn that regression of data for Y1 versus Y3a1 a2 gives a slope equal to 1.385 with lower and upper 95% confidence intervals equal to 1.22 and 1.55. Does this ‘ test ’ confirm our theory, or is it possible that an alternative theory positing a1=2/3 and a2=2 such that a1a2 is predicted to equal 4/3 is correct ? Clearly, 4/3=1.333 falls well within the empirically determined 95 % confidence intervals of the data for Y1 versus Y3 a1 a2 , whereas both theories predict that a2=2. Thus, the only remaining test to determine which of the two theories is correct is whether the slope of Y1 versus Y2 a1 is numerically more in accord with a1=3/4 or 2/3. Accordingly, the multiple predictions of the two theories are reduced to one in terms of statistical testing. (5 ) Different ‘ first principles’ can yield the same predictions Currently, the most comprehensive allometric theory in biology is that of West, Brown & Enquist (1997, 1999, 2000 ; see also Brown, West & Enquist, 2000; Enquist, 2002). The WBE theory attempts to explain an extraordinarily broad spectrum of scaling relationships, ranging from physiological processes to community-level and macroecological phenomena. Importantly, it rests on three assumptions relating to the exchange, distribution and delivery networks for energy and mass transfer in biological systems. These networks are argued to (1) branch three-dimensionally throughout biological structures (within cells or entire organisms), (2) have terminal units that do not vary in size 887 within organisms (e.g. capillaries and peripheral xylem elements), and (3) minimize the total resistance (and thus the energy and time) involved in the distribution and delivery of resources. In simplistic terms, the WBE theory states that organisms, whether unicellular or multicellular, have internal networks with fractal-like architectures that have evolved through natural selection to minimize the energy and time required to absorb, distribute, and deliver resources internally. These assertions mathematically obtain the prediction that biological surface areas scale as the 3/4 power of body volumes and that many other size-correlated biological phenomena obey 1/4 (or its multiples) power ‘ rules. ’ In this way, the historically classic 3/4 scaling relationship observed by Kleiber and many others receives a formal (mathematical and conceptual) explanation. Support for the WBE theory, however, is currently circumstantial. True, many allometric trends seem to have scaling exponents of 1/4 or multiples thereof. However, from a purely logical perspective, the verification of any theory requires the validation of its basic assumptions and not its predictions if for no other reason than that alternative theories may obtain the same predictions starting with different ‘first principle ’ assumptions. The allometric derivations presented here are a case in point. Each set of derivations is based in part or whole on the empirical observation that annual plant growth rates appear to scale as the 3/4 power of body mass. From this basic scaling ‘ rule, ’ a variety of other allometric relationships emerge, each of which is governed by quarter-power scaling exponents. It is nevertheless illogical to interpret this as evidence supporting the WBE theory because any set of derivations that employs the 3/4 scaling relationship between growth and body mass (for whatever reason) may predict many of the same scaling relations as those presented here. The argument that no ‘ first principles ’ theory currently exists other than the WBE theory is both logically irrelevant and untrue (see Weibel, 2000; Darveau et al., 2002). Because its basic assumptions have yet to be subjected to fastidious and extensive tests and because ad hoc explanations can always be advanced to explain away discrepancies between observed and predicted scaling exponents, it is premature to pass judgment on the WBE theory. Certainly, it has reinvigorated the study of biological allometry in general and the study of plant allometry in particular. What can be said with more certainty is that there are size-correlated trends that span many orders of magnitude in body size across virtually every plant lineage and habitat. That these trends exist at all suggests that they reflect very fundamental properties of living things that demand a mechanistic explanation. XI. CONCLUSIONS (1) Two allometric and empirical trends receive strong statistical support based on broad interspecific comparisons : annual growth in biomass at the level of an individual plant GT scales as the 3/4 power of body mass MT and Karl J. Niklas 888 isometrically with respect to the ability of an individual plant (unicellular or multicellular) to harvest light H, i.e. GT ! M3T/4 ! H. (2) With the aid of only a few assumptions concerning stem and root hydraulics (e.g. standing leaf biomass ML scales as the square of basal stem diameter DS), these two trends provide a basis to analytically derive a spectrum of hypotheses concerning size-correlated phenomena, each of which is seen to receive strong statistical support, e.g., standing leaf biomass scales as the 3/4 power of standing stem (or root) biomass ; annual growth in leaf biomass scales isometrically with respect to annual growth in stem (or root) biomass ; annual reproductive biomass scales as a speciesspecific complex function of how annual growth is partitioned among leaf, stem, and root biomass ; total body biomass scales as the x4/3 power of plant density. (3) Although the general allometric theory emerging from these derivations is far-reaching and statistically robust, the theoretical basis for the allometric trends GT ! MT3/4 ! H remains problematic because any theory that starts with the assumption that body surface area scales as the 3/4 power of body volume (mass) will obtain the same predictions regarding such features as standing leaf, stem, and root biomass allocation patterns. (4) Additionally, a number of statistical and mathematical issues regarding allometric analyses (e.g., the choice of regression analysis, and converting the numerical values of scaling exponents into fractions) must be carefully explored before rival allometric theories can be resolved using large data sets. XII. LIST OF SYMBOLS a, scaling exponent (slope of log–log linear regression curve). aRMA, scaling exponent (slope of log–log linear reduced major axis regression curve). b, allometric constant (Y-intercept of log–log linear regression curve). bRMA, allometric constant (Y-intercept of log–log linear reduced major axis regression curve). ai, unit area occupied by an individual in a community. AT, total area occupied by all individuals in a community. DS, DR, average diameter of stem and root, respectively. GL, GS, GR, GP, annual growth in leaf, stem, root, and reproductive biomass plantx1, respectively. GT, annual growth in biomass plantx1. k, a numerical constant. LS, LR, average length of stem and root, respectively. MT, total biomass plantx1. ML, MS, MR, MSH, standing leaf, stem, root, and shoot (leaf and stem) biomass plantx1, respectively. MP, standing reproductive biomass plantx1. M̂L, M̂S, M̂R, M̂SH, standing leaf, stem, root, and shoot (leaf and stem) biomass communityx1, respectively. N, plant density (number of plants occupying AT,). r, correlation coefficient. rS, rR, bulk tissue density of stem and root, respectively. t, time. X, Y, independent and dependent variables, respectively. y1, y2, numerical values of interdependent variables Y1, Y2. Y1, Y2, interdependent variables. XIII. ACKNOWLEDGEMENTS Many of the concepts and analyses presented here have been published together with Brian Enquist, Jeremy Midgley, and Richard Rand, whom I gratefully acknowledge. I am also grateful to two anonymous reviewers who provided valuable comments on how to improve the organization of this article. This research has been supported by funds provided by the College of Agricultural and Life Sciences, Cornell University. XIV. REFERENCES BANAVAR, J. R., MARITAN, A. & RINALDO, A. (1999). Size and form in efficient transportation networks. Nature 399, 130–132. BLUM, J. J. (1977). On the geometry of four-dimensions and the relationship between metabolism and body mass. Journal of Theoretical Biology 64, 599–601. BOND-LAMBERTY, B., WANG, C. & GOWER, S. T. (2001). Aboveground and belowground biomass and sapwood area allometric equations for six boreal tree species of northern Manitoba. Canadian Journal of Forestry Research 32, 1441–1450. BRODY, S. (1945). Bioenergetics and Growth. Reinhold, New York. BROWN, J. H. & WEST, G. B. (2000). Scaling in Biology. Oxford University Press, New York. BROWN, J. H., WEST, G. B. & ENQUIST, B. J. (2000). Scaling in biology : patterns and processes, causes and consequences. In Scaling in Biology (eds. J. H. Brown and G. W. West), pp. 1–24. Oxford University Press, New York. CALDER, W. A. III. (1984). Size, Function, and Life History. Harvard University Press, Cambridge. CANNELL, M. G. R. (1982). World Forest Biomass and Primary Production Data. Academic Press, London. CARLQUIST, S. (1975). Ecological Strategies of Xylem Evolution. University of California Press, Berkeley. CHAPPELL, R. (1989). Fitting bent lines to data, with applications to allometry. Journal of Theoretical Biology 138, 235–256. DARVEAU, C. A., SUAREZ, R. K., ANDREWS, R. D. & HOCHACHKA, P. W. (2002). Allometric cascade as a unifying principle of body mass effects on metabolism. Nature 417, 166–170. DODDS, P. S., ROTHMAN, D. H. & WEITZ, J. S. (2001). Re-examination of the ‘‘ 3/4-law’’ of metabolism. Journal of Theoretical Biology 209, 9–27. ENQUIST, B. J. (2002). Universal scaling in tree and vascular plant allometry : toward a general quantitative theory linking plant form and function from cells to ecosystems. Tree Physiology 22, 1045–1064. ENQUIST, B. J. & NIKLAS, K. J. (2001). Invariant scaling relations across tree-dominated communities. Nature 410, 655–660. ENQUIST, B. J. & NIKLAS, K. J. (2002). Global allocation rules for patterns of biomass partitioning across seed plants. Science 295, 1517–1520. FRANCO, M. & KELLY, C. K. (1998). The interspecific mass-density relationship and plant geometry. Proceedings of the National Academy of Sciences (USA) 95, 7830–7835. GARTNER, B. L. (1995). Patterns of xylem variation within a tree and their hydraulic and mechanical consequences. In Plant Plant allometry Stems (ed. B. L. Gartner), pp. 125–150. Academic Press, San Diego. GENARD, M., FISHMAN, S., VERCAMBRE, G., HUGUET, J. G., BUSSI, C., BESSET, J. & HABIB, R. (2001). A biophysical analysis of stem and root diameter variation in woody plants. Plant Physiology 126, 188–202. GOULD, S. J. (1966). Allometry and size in ontogeny and phylogeny. Biological Reviews 41, 587–640. GRAY, B. F. (1981). On the ‘ surface law ’ and basal metabolic rate. Journal of Theoretical Biology 93, 757–767. HARVEY, P. H. (1982). On rethinking allometry. Journal of Theoretical Biology 95, 37–41. HARVEY, P. H. & MACE, G. M. (1982). Comparisons between taxa and adaptive trends : problems of methodology. In Current Problems in Sociobiology (eds. King’s College Sociobiology Group), pp. 343–361. Cambridge University Press, Cambridge. HEMMINGSEN, A. M. (1960). Energy metabolism as related to body size and respiratory surfaces, and its evolution. Reports of the Steno Memorial Hospital and Nordisk Insulin Laboratorium 9, 6–110. HOLBROOK, N. M. (1995). Stem water storage. In Plant Stems (ed. B. L. Gartner), pp. 151–176. Academic Press, San Diego. HUXLEY, J. S. (1924). Constant differential growth-ratios and their significance. Nature 114, 895. HUXLEY, J. S. (1932). Problems of Relative Growth. Methuen & Co., Ltd. London. JOLICOEUR, P. & MOSIMANN, J. E. (1968). Intervalles de confiance pour la pente de l’axe majeur d’une distribution normale bidimensionnelle. Biométrie-Praximétrie 9, 121–140. KERMACK, K. A. & HALDANE, J. B. S. (1950). Organic correlation and allometry. Biometrika 37, 30–41. KLEIBER, M. (1947). Body size and metabolic rate. Physiological Review 27, 511–541. KÖRNER, C. (1994). Biomass fractional in plants : a reconsideration of definitions on plant functions. In A Whole Plant Perspective on Carbon-Nitrogen Interactions (eds. J. Roy and E. Garnier), pp. 173–185. SPB Academic Publishing, The Hague. LABARBERA, M. (1986). The evolution and ecology of body size. In Patterns and Processes in the History of Life (eds. D. M. Raup and D. Jablonski), pp. 69–98. Springer-Verlag, Berlin. LABARBERA, M. (1989). Analyzing body size as a factor in ecology and evolution. Annual Review of Ecology and Systematics 20, 97–117. MAKKONEN, K. & HELMISSAR, H. S. (2001). Fine root biomass and production in Scots pine stands relative to stand age. Tree Physiology 21, 193–198. MCARDLE, B. H. (1988). The structural relationship : regression in biology. Canadian Journal of Zoology 66, 2329–2339. MCMAHON, T. A. (1973). Size and shape in biology. Science 179, 1201–1204. MCMAHON, T. A. (1980). Food habits, energetics, and the population biology of mammals. American Naturalist 116, 106–124. MCMAHON, T. A. & BONNER, J. T. (1983). On Size and Life. Scientific American Books, New York. NIKLAS, K. J. (1994 a). Morphological evolution through complex domains of fitness. Proceedings of the National Academy of Sciences (USA) 91, 6772–6779. NIKLAS, K. J. (1994 b). Plant Allometry. University of Chicago Press, Chicago. NIKLAS, K. J. (2002). On the allometry of biomass partitioning and light harvesting for plants with leafless stems. Journal of Theoretical Biology 217, 47–52. 889 NIKLAS, K. J. (2003). Reexamination of a canonical model for plant organ biomass partitioning. American Journal of Botany 90, 250–254. NIKLAS, K. J. & ENQUIST, B. J. (2001). Invariant scaling relationships for interspecific plant biomass production rates and body size. Proceedings of the National Academy of Sciences (USA) 98, 2922–2927. NIKLAS, K. J. & ENQUIST, B. J. (2002 a). Canonical rules for plant organ biomass partitioning and growth allocation. American Journal of Botany 89, 812–819. NIKLAS, K. J. & ENQUIST, B. J. (2002 b). On the vegetative biomass partitioning of seed plant leaves, stems, and roots. American Naturalist 159, 482–497. NIKLAS, K. J. & ENQUIST, B. J. (2003). An allometric model for seed plant reproduction. Evolutionary Ecology Research 5, 79–58. NIKLAS, K. J., MOLINA-FREANER, F., TINOCO-OJAMGUREN, C. & PAOLILLO, D. J. JR. (2002). The biomechanics of Pachycereus pringlei root systems. American Journal of Botany 89, 12–21. NIKLAS, K. J., MIDGLEY, J. J. & ENQUIST, B. J. (2003 a). A general model for mass-growth-density relations across tree-dominated communities. Evolutionary Ecological Research 5, 459–468. NIKLAS, K. J., MIDGLEY, J. J. & RAND, R. H. (2003 b). Tree size distributions, plant density, age, and community disturbance. Ecology Letters 6, 405–411. PETERS, R. H. (1983). The Ecological Implications of Body Size. Cambridge University Press, Cambridge. PFANZ, H., ASCHAN, G., LANGENFELD-HEYSER, R., WITTMAN, C. & LOOSE, M. (2002). Ecology and ecophysiology of tree stems : corticular and wood photosynthesis. Naturwissenschaften 89, 147–162. PROTHERO, J. (1986). Methodological aspects of scaling in biology. Journal of Theoretical Biology 118, 259–286. RAYNER, J. M. V. (1985). Linear relations in biomechanics : the statistics of scaling functions. Journal of Zoology 206, 415–439. SCHMIDT-NIELSON, K. (1984). Scaling, Why is Animal Size so Important ? Cambridge University Press, Cambridge. SEIM, E. (1983). On rethinking allometry : which regression model to use ? Journal of Theoretical Biology 104, 161–168. SOKAL, R. R. & ROHLF, F. J. (1981). Biometry, 2nd edn. W. H. Freedman and Co., New York. SMITH, R. J. (1980). Rethinking allometry. Journal of Theoretical Biology 87, 97–111. THOMPSON, D’A. W. (1942). On Growth and Form, 2nd edn. Cambridge University Press, Cambridge. WEIBEL, E. R. (2000). Synmmorphosis. Harvard University Press, Boston. WEIBEL, E. R. (2002). Physiology – the pitfalls of power laws. Nature 417, 131–132. WEST, G. B., BROWN, J. H. & ENQUIST, B. J. (1997). A general model for the origin of allometry scaling laws in biology. Science 276, 122–126. WEST, G. B., BROWN, J. H. & ENQUIST, B. J. (1999). The fourth dimension of life : fractal geometry and allometric scaling of organisms. Science 284, 167–169. WEST, G. B., BROWN, J. H. & ENQUIST, B. J. (2000). The origin of universal scaling laws in biology. In Scaling in Biology (eds. J. H. Brown and G. W. West), pp. 87–112. Oxford University Press, New York. ZIMMERMANN, M. H. (1983). Xylem Structure and the Ascent of Sap. Springer-Verlag, Berlin.