An Efficient Algorithm for Computing Non
advertisement

The 32nd Workshop on Combinatorial Mathematics and Computation Theory
An Efficient Algorithm for Computing Non-overlapping Inversion
and Transposition Distance
Toan Thang Ta, Cheng-Yao Lin and Chin Lung Lu
Department of Computer Science
National Tsing Hua University, Hsinchu 30013, Taiwan
{toanthanghy, begoingto0830}@gmail.com, cllu@cs.nthu.edu.tw
given pattern with non-overlapping reversals,
where n is the length of the text and m is the
length of the pattern. In this problem, two
equal-length strings are said to have a match with
non-overlapping reversals if one string can be
transformed into the other using any finite
sequence of non-overlapping reversals. It should
be noted that the number of the used
non-overlapping reversals in the algorithm
proposed by Cantone et al. [2] is not required to be
less than or equal to a fixed non-negative integer.
In [2], Cantone et al. also presented another
algorithm whose average-case time complexity is
ܱሺ݊ሻ. Cantone et al. [3] studied the same problem
by considering both non-overlapping reversals and
transpositions, where they called transpositions as
translocations and the lengths of two exchanged
adjacent fragments are constrained to be
equivalent. They finally designed an algorithm to
solve this problem in ܱሺ݊݉ଶ ሻ time and ܱሺ݉ଶ ሻ
space. For the above problem, Grabowski et at. [6]
gave another algorithm whose worst-case time and
space are ܱሺ݊݉ଶ ሻ and ܱሺ݉ሻ , respectively.
Moreover, they proved that their algorithm has an
ܱሺ݊ሻ average time complexity. Recently, Huang
et al. [7] studied the above approximate string
matching problem under non-overlapping
reversals by further restricting the number of the
used reversals not to exceed a given positive
integer ݇. They proposed a dynamic programming
algorithm to solve this problem in ܱሺ݊݉ଶ ሻ time
and ܱሺ݉ଶ ሻ space.
In this work, we are interested in the
computation of the mutation distance between two
strings of the same length under non-overlapping
inversions
and
transpositions
(i.e.,
non-overlapping inversion and transposition
distance), where the lengths of two adjacent and
non-overlapping fragments exchanged by a
transposition can be different. For this problem,
we devise an algorithm whose time and space
complexities are ܱሺ݊ଷ ሻ and ܱሺ݊ଶ ሻ, respectively,
where ݊ is the length of two input strings. The
rest of the paper is organized as follows. In
Section 2, we provide some notations that are
helpful when we present our algorithm later. Next,
Abstract
Given two strings of the same length ݊, the
non-overlapping inversion and transposition
distance (also called mutation distance) between
them is defined as the minimum number of
non-overlapping inversion and transposition
operations used to transform one string into the
other. In this study, we present an ܱሺ݊ଷ ሻ time and
ܱሺ݊ଶ ሻ space algorithm to compute the mutation
distance of two input strings.
1
Introduction
The dissimilarity of two strings is usually
measured by the so-called edit distance, which is
defined as the minimum number of edit operations
necessary to convert one string into the other. The
commonly used edit operations are character
insertions, deletions and substitutions. In
biological application, the aforementioned edit
operations correspond to point mutations of DNA
sequences (i.e., mutations at the level of individual
nucleotides). From evolutionary point of view,
however, DNA sequences may evolve by
large-scale mutations (also called rearrangements,
i.e., mutations at the level of sequence fragments)
[8], such as inversions (replacing a fragment of
DNA sequence by its reverse complement) and
transpositions (i.e., moving a fragment of DNA
sequence from one location to another or,
equivalently, exchanging two adjacent and
non-overlapping fragments on DNA sequence).
Note that a large-scale mutation that replaces a
fragment of DNA sequence only by its reverse
(without complement) is called a reversal. Based
on large-scale mutation operations, the
dissimilarity (or mutation distance) between two
strings can be defined to be the minimum number
of large-scale mutation operations used to
transform one string to the other. Cantone et al. [2]
introduced an ܱሺ݊݉ሻ time and ܱሺ݉ଶ ሻ space
algorithm to solve an approximate string matching
problem with non-overlapping reversals, which is
to find all locations of a given text that match a
55
The 32nd Workshop on Combinatorial Mathematics and Computation Theory
we develop the main algorithm and also analyze
its time and space complexities in Section 3.
Finally, we have a brief conclusion in Section 4.
mutations that converts ݔinto ݕ, then ݉݀ሺݔǡ ݕሻ
is infinite. Formally,
2
ቄ
݉݀ሺݔǡ ݕሻ ൌ
Preliminaries
Let ݔbe a string of length ݊ over a finite
alphabet 6. A character at position ݅ of x is
represented with ݔ , where ͳ ݅ ݊ . A
substring of ݔfrom position ݅ to ݆ is indicated
as ݔǡ , i.e., ݔǡ ൌ ݔ ݔାଵ ǥ ݔ , for ͳ ݅ ݆ ݊.
In biological sequences, ȭ ൌ ሼܣǡ ܥǡ ܩǡ ܶሽ , in
which ܣ- ܶ and ܥ- ܩare considered as
complementary base pairs. We use ߠሺݔሻ to
denote an inversion operation acting on a string ݔ,
resulting in a reverse and complement of ݔ. For
example, ߠሺܣሻ ൌ ܶ , ߠሺܶሻ ൌ ܣ, ߠሺܩሻ ൌ ܥ,
ߠሺܥሻ ൌ ܩand ߠሺܣܩܥሻ ൌ ܶܩܥ. In addition, we
utilize ߬ሺݒݑሻ ൌ ݑݒto represent a transposition
operation to exchange two non-empty strings ݑ
and ݒ. Note that the lengths of ݑand ݒare
required to be identical in some previous works [3,
5, 6], but they may be different in this study. For
convenience, we call ߠ and ߬ as mutation
operations. We also let ߠǡ ሺݔሻ ൌ ߠሺݔǡ ሻ for ͳ
݅ ݆ ݊ and ߬ǡǡ ሺݔሻ ൌ ݔǡ ݔǡିଵ for ͳ ݅ ൏
݇ ݆ ݊, where ሾ݅ǡ ݆ሿ is called a mutation range
for ߠǡ and ߬ǡǡ and ݇ is called as a cut point
in ߬ǡǡ .
For an integer ͳ ݐ ݊ , we say that a
mutation operation ߠǡ or ߬ǡǡ covers ݐif ݅
ݐ ݆. Given two mutation operations, they are
non-overlapping if the intersection of their
mutation ranges are empty. In this study, we are
only interested in non-overlapping mutation
operations. Consider a set 4 of non-overlapping
mutation operations and a string ݔ, let 4ሺݔሻ be
the resulting string after consecutively applying
mutation operations in 4 on ݔ. For example,
suppose that 4 ൌ ൛߬ଵǡଷǡଶ ǡ ߠହǡହ ൟ and ݔൌ ܶܥܣܩܣ.
Then we have 4ሺݔሻ ൌ ܩܣܶܩܣ.
ሼȁ4ȁ 4ሺݔሻ ൌ ݕሽ
f
4
4ሺݔሻ ൌ ݕ
For example, consider ݔൌ ܶ ܥܣܩܣand ݕൌ
ܶ ܩܥܣܣ. Then there are only two sets of
non-overlapping mutation operations 4ଵ ൌ
൛ߠଵǡଶ ǡ ߬ଷǡହǡସ ൟ and 4ଶ ൌ ൛߬ଷǡହǡସ ൟ such that 4ଵ ሺݔሻ ൌ
ݕand 4ଶ ሺݔሻ ൌ ݕ. Thus, ݉݀ሺݔǡ ݕሻ = ȁ4ଶ ȁ ൌ ͳ.
3
The algorithm
Basically, a transposition (respectively,
inversion) operation acting on a string ݔcan be
considered as a permutation of characters in ݔ
(respectively, complement of )ݔ. From this view
point, a mutation operation actually comprises
several sub-operations, called as mutation
fragments, each of which is denoted either by a
tuple ሺ݅ǡ ݆ǡ ݔ ሻ or ሺ݅ǡ ݆ǡ ߠሺݔ ሻሻ . The mutation
fragment ሺ݅ǡ ݆ǡ ݔ ሻ (respectively, ሺ݅ǡ ݆ǡ ߠሺݔ ሻሻ )
means that ݔ (respectively, complement of ݔ ) is
moved into the position ݅ in the resulting string
obtained when applying the mutation operation on
ݔ. For convenience, we arrange all possible
mutation fragments in a three-dimensional table
ܯ௫ ሾʹǡ ݊ǡ ݊ሿ, which is called mutation table of ݔ,
as follows.
x ܯ௫ ሾͳǡ ݅ǡ ݆ሿ ൌ ሺ݅ǡ ݆ǡ ߠሺݔ ሻሻfor ݅ǡ ݆ ൌ ͳǡ ʹǡ ǥ ǡ ݊.
x ܯ௫ ሾʹǡ ݅ǡ ݆ሿ ൌ ሺ݅ǡ ݆ǡ ݔ ሻ for ݅ǡ ݆ ൌ ͳǡ ʹǡ ǥ ǡ ݊.
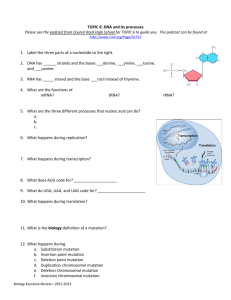
For example, let ݔൌ ܶ ܥܣܩܣ. Then its
mutation table is shown in Figure 1.
When an inversion operation ߠǡ applies on a
string ݔ, we can decompose it into ሺ݆ െ ݅ ͳሻ
mutation fragments ࣠ሺߠǡ ǡ ݔǡ ݐሻ for ݅ ݐ ݆ ,
where ࣠ሺߠǡ ǡ ݔǡ ݐሻ ൌ ሺݐǡ ݅ ݆ െ ݐ, ߠሺݔାି௧ ሻሻ .
Similarly, a transposition operation ߬ǡǡ can be
also decomposed into ሺ݆ െ ݅ ͳሻ mutation
fragments ࣠ሺ߬ǡǡ ǡ ݔǡ ݐሻ for ݅ ݐ ݆ , where if
݅ ݐ ݅ ݆ െ ݇ , then ࣠ሺ߬ǡǡ ǡ ݔǡ ݐሻ ൌ ሺݐǡ ݇
ݐെ ݅ǡ ݔା௧ି ሻ; otherwise (i.e., if ݅ ݆ െ ݇ ൏ ݐ
݆ ),
then
࣠ሺ߬ǡǡ ǡ ݔǡ ݐሻ ൌ ሺݐǡ ݐെ ݆ ݇ െ
ͳ,ݔ௧ିାିଵ ሻ. For the sake of succinctness, let
߬ǡǡ ሺݔǡ ͳሻ ൌ ሼሺݐǡ ݇ ݐെ ݅ǡ ݔା௧ି ሻ ݅ ݐ ݅
݆ െ ݇ሽ
and
߬ǡǡ ሺݔǡ ʹሻ ൌ ൛ሺݐǡ ݐെ ݆ ݇ െ
ͳ,ݔ௧ିାିଵ ሻ ݅ ݆ െ ݇ ൏ ݐ ݆ൟ.
Definition 1. (Non-overlapping inversion and
transposition distance) Given two strings ݔand
ݕof the same length, the non-overlapping
inversion and transposition distance (simply called
mutation distance) between ݔand ݕ, denoted by
݉݀ሺݔǡ ݕሻ, is defined as the minimum number of
non-overlapping inversion and transposition
operations used to transform ݔinto ݕ. If there
does not exist any set of non-overlapping
56
The 32nd Workshop on Combinatorial Mathematics and Computation Theory
ܯ௫ ሾͳǡ ݅ǡ ݆ሿ
݅ ൌ 1
2
3
4
5
݆ ൌ 1
ሺͳǡͳǡ ܣሻ
ሺʹǡͳǡ ܣሻ
ሺ͵ǡͳǡ ܣሻ
ሺͶǡͳǡ ܣሻ
ሺͷǡͳǡ ܣሻ
2
ሺͳǡʹǡ ܶሻ
ሺʹǡʹǡ ܶሻ
ሺ͵ǡʹǡ ܶሻ
ሺͶǡʹǡ ܶሻ
ሺͷǡʹǡ ܶሻ
3
ሺͳǡ͵ǡ ܥሻ
ሺʹǡ͵ǡ ܥሻ
ሺ͵ǡ͵ǡ ܥሻ
ሺͶǡ͵ǡ ܥሻ
ሺͷǡ͵ǡ ܥሻ
4
ሺͳǡͶǡ ܶሻ
ሺʹǡͶǡ ܶሻ
ሺ͵ǡͶǡ ܶሻ
ሺͶǡͶǡ ܶሻ
ሺͷǡͶǡ ܶሻ
5
ሺͳǡͷǡ ܩሻ
ሺʹǡͷǡ ܩሻ
ሺ͵ǡͷǡ ܩሻ
ሺͶǡͷǡ ܩሻ
ሺͷǡͷǡ ܩሻ
ܯ௫ ሾʹǡ ݅ǡ ݆ሿ
݅ ൌ 1
2
3
4
5
݆ ൌ 1
ሺͳǡͳǡ ܶሻ
ሺʹǡͳǡ ܶሻ
ሺ͵ǡͳǡ ܶሻ
ሺͶǡͳǡ ܶሻ
ሺͷǡͳǡ ܶሻ
2
ሺͳǡʹǡ ܣሻ
ሺʹǡʹǡ ܣሻ
ሺ͵ǡʹǡ ܣሻ
ሺͶǡʹǡ ܣሻ
ሺͷǡʹǡ ܣሻ
3
ሺͳǡ͵ǡ ܩሻ
ሺʹǡ͵ǡ ܩሻ
ሺ͵ǡ͵ǡ ܩሻ
ሺͶǡ͵ǡ ܩሻ
ሺͷǡ͵ǡ ܩሻ
4
ሺͳǡͶǡ ܣሻ
ሺʹǡͶǡ ܣሻ
ሺ͵ǡͶǡ ܣሻ
ሺͶǡͶǡ ܣሻ
ሺͷǡͶǡ ܣሻ
5
ሺͳǡͷǡ ܥሻ
ሺʹǡͷǡ ܥሻ
ሺ͵ǡͷǡ ܥሻ
ሺͶǡͷǡ ܥሻ
ሺͷǡͷǡ ܥሻ
Figure 1. A mutation table ܯ௫ for a given string ݔൌ ܶܥܣܩܣ, where the column is indexed by ݅ and the row
by ݆. Shaded cells respectively represent the inversion ߠଵǡଷ ሺݔሻ on ܯ௫ ሾͳǡ ݅ǡ ݆ሿ and the transposition operation
߬ଵǡହǡସ ሺݔሻ on ܯ௫ ሾʹǡ ݅ǡ ݆ሿǤ
κሺܨሻ ൌ ߪ . If a sequence ܵ ൌ ܨۃଵ ǡ ܨଶ ǡ ǥ ǡ ܨ ۄ
consists of ݉ mutation fragments ( ݉ is also
considered as the length of ܵ) with κሺܨ ሻ ൌ ߪ
for ͳ ݅ ݉, then we say that ܵ yields a string
ߪଵ ߪଶ ǥ ߪ , which is further written as κሺSሻ ൌ
ߪଵ ߪଶ ǥ ߪ . A subsequence ܲ containing first ݐ
elements of ܵ, i.e., ܲ ൌ ܨۃଵ ǡ ܨଶ ǡ ǥ ǡ ܨ௧ ۄ, is called a
prefix of ܵ and denoted by ܲ ٌ ܵ, where ͳ
ݐ ݉.
The aforementioned decomposition can be
extended to apply on a set 4 of non-overlapping
mutation operations. That is, when 4 acts on a
string ݔ, it can be decomposed into a sequence
࣠ሺ4ǡ ݔሻ ൌ ܨۃଵ ǡ ܨଶ ǡ ǥ ǡ ܨ ۄof mutation fragments
by the following formula: For ݐൌ ͳǡ ʹǡ ǥ ǡ ݊,
ߠǡ א4
ݐ
࣠ሺߠǡ ǡ ݔǡ ݐሻ
ܨ௧ ൌ ቐ࣠ሺ߬ǡǡ ǡ ݔǡ ݐሻ ߬ǡǡ א4
ݐ
ሺݐǡ ݐǡ ݔ௧ ሻ
Definition 2. (Agreed sequence) A sequence ܵ ൌ
ܨۃଵ ǡ ܨଶ ǡ ǥ ǡ ܨ ۄof ݉ mutation fragments is an
agreed sequence if there exists a set 4 of
non-overlapping mutation operations such that
ܵ ٌ ࣠ሺ4ǡ ݔሻ, where ݉ ݊.
Given a mutation operation ߠǡ or ߬ǡǡ , we
call ݅ and ݆ as the left end point and right end
point of the mutation operation, respectively. We
also say that this mutation operation (i.e., ߠǡ or
߬ǡǡ ) intersects with a range ሾܽǡ ܾሿ if the
intersection of ሾ݅ǡ ݆ሿ and ሾܽǡ ܾሿ is non-empty.
The number of mutation operations in 4 that
intersects with the range ሾͳǡ ݉ሿ where ݉ ݊ is
denoted as ߩሺ4ǡ ݉ሻ . For example, if 4 ൌ
൛ߠଵǡଶ ǡ ߬ସǡǡହ ǡ ߠǡଽ ൟ, then ߩሺ4ǡ Ͷሻ ൌ ʹ. Suppose that
a mutation fragment ܨൌ ሺ݅ǡ ݆ǡ ߪሻ is in ࣠ሺ4ǡ ݔሻ
for some set 4 of non-overlapping mutation
operations. Then ܨis said to be covered by a
mutation operation ߠǡ or ߬ǡǡ in 4 if ܨൌ
࣠ሺߠǡ ǡ ݔǡ ݅ሻ or ܨൌ ࣠ሺ߬ǡǡ ǡ ݔǡ ݅ሻ . If ܨis not
For instance, given 4 ൌ ൛߬ଵǡଶǡଶ ǡ ߠସǡହ ൟ and ݔൌ
ܶ ܥܣܩܣ, we have ࣠ሺ4ǡ ݔሻ ൌ ۦሺͳǡʹǡ ܣሻǡ ሺʹǡͳǡ ܶሻǡġ
ሺ͵ǡ͵ǡ ܩሻǡ ሺͶǡͷǡ ܩሻǡ ሺͷǡͶǡ ܶሻۧį
Observation 1. Given a string x and its mutation
table ܯ௫ , the result of an inversion ߠǡ ሺݔሻ can
be obtained by concatenating ሺ݆ െ ݅ ͳሻ
mutation fragments starting at ܯ௫ ሾͳǡ ݅ǡ ݆ሿ and
continuing to move in the anti-diagonal direction
to ܯ௫ ሾͳǡ ݆ǡ ݅ሿ . Moreover, the result of a
is
obtained
by
transposition
߬ǡǡ ሺݔሻ
concatenating the mutation fragments on the
following two paths, one starting at ܯ௫ ሾʹǡ ݅ǡ ݇ሿ
and continuing to move in the diagonal direction
to ܯ௫ ሾʹǡ ݅ ݆ െ ݇ǡ ݆ሿ and the other beginning at
ܯ௫ ሾʹǡ ݅ ݆ െ ݇ ͳǡ ݅ሿ and also continuing to
move in the diagonal direction to ܯ௫ ሾʹǡ ݆ǡ ݇ െ ͳሿ.
For a mutation fragment ܨൌ ሺ݅ǡ ݆ǡ ߪሻ, we say
that ܨyields the character ߪ , denoted by
57
The 32nd Workshop on Combinatorial Mathematics and Computation Theory
covered by any mutation operation in 4, that is
ܨൌ ሺ݅ǡ ݅ǡ ݔ ሻ, then we can consider that ܨis still
covered by a virtual mutation operation. We use
ࣦሺ4ǡ ݔǡ ܨሻ and ࣬ሺ4ǡ ݔǡ ܨሻ to denote the left and
right end points of a mutation operation in 4
respectively that covers ܨ. Formally,
length ݅ ͳ , which can be obtained from all
previously created legal sequences of length ݅. At
the end, if there exists a legal sequence of length
݊, then the algorithm returns the minimum number
of the used mutation operations among all legal
sequences of length ݊. Otherwise, the algorithm
returns infinity.
To distinguish all the legal sequences of length
݅ , we define three sets of extended mutation
fragments ܫ௫ , ܸ௫ and ܶ௫ of x as follows:
x ܫ௫ ൌ ቄ൬݃ǡ ݀ǡ ቀ݅ǡ ݆ǡ ߠ൫ݔ ൯ቁ൰ቅ for Ͳ ݃ ݊ െ ͳǡ
Ͳ ݀ ݊ and ݆ ൌ ͳǡ ʹǡ ǥ ǡ ݊.
x ܸ௫ ൌ ൛൫Ͳǡ ݀ǡ ሺ݅ǡ ݅ǡ ݔ ሻ൯ൟ for Ͳ ݀ ݊.
x ܶ௫ ൌ ൛൫ݏǡ ݀ǡ1ǡ ሺ݅ǡ ݆ǡ ݔ ሻ൯ൟ ൛൫݃ǡ ݀ǡ ʹǡ ሺ݅ǡ ݆ǡ ݔ ሻ൯ൟ
for ͳ ݏ ݊ െ ͳ, Ͳ ݃ ݊ െ ͳ, Ͳ ݀ ݊
and ݆ ൌ ͳǡ ʹǡ ǥ ǡ ݊.
Actually, each extended mutation fragment
contains the last mutation fragment ܨof some
agreed sequence ܵ of length ݅ with extra guiding
information ݃ǡ ݀ and ݏ. The value of ݃ is the
number of steps that are required to move towards
the anti-diagonal or diagonal direction to complete
a mutation operation and ݀ indicates the number
of the currently used mutation operations in ܵ. If
the last mutation fragment ܨis covered by a
transposition operation, say ݉௫ , and א ܨ
݉௫ ሺݔǡ ͳሻ, then ݏdenotes the left end point of ݉௫ .
An extended mutation fragment is further called
an ending fragment (respectively, continuing
fragment) if the value of its first component is
zero (respectively, non-zero).
Next, we represent each legal sequence by an
extended fragment as follows. Given a legal
sequence ܵ ൌ ܨۃଵ ǡ ܨଶ ǡ ǥ ǡ ܨ ۄthat consists of ݅
mutation fragments of ݔ, let 4 be any set of
non-overlapping mutation operations such that
ܵ ٌ ࣠ሺ4ǡ ݔሻ. We further let ݉௫ be the mutation
operation in 4 that covers ܨ . Note that ݉௫ may
be a virtual mutation operation. We create an
extended fragment ݁ by the following cases (each
case is also called as a type of extended mutation
fragment):
Case 1 (type )ܫ: ݉௫ is an inversion operation.
Then ݁ ൌ ሺ࣬ሺ4ǡ ݔǡ ܨ ሻ െ ݅ǡ ߩሺ4ǡ ݅ሻǡ ܨ ሻ.
Case 2 (type ܸ ): ݉௫ is a virtual mutation
operation. Then ݁ ൌ ሺͲǡ ߩሺ4ǡ ݅ሻǡ ܨ ሻ.
Case 3 (type ܨሻǣ ݉௫ is a transposition operation
and ܨ ݉ א௫ ሺݔǡ ͳሻǤ Then ݁ ൌ ሺࣦሺ4ǡ ݔǡ ܨ ሻǡ
ߩሺ4ǡ ݅ሻǡ ͳǡ ܨ ሻ.
Case 4 (type ܵሻ: ݉௫ is a transposition operation
and ܨ ݉ א௫ ሺݔǡ ʹሻǤ Then ݁ ൌ ሺ࣬ሺ4ǡ ݔǡ ܨ ሻ െ
݅ǡ ߩሺ4ǡ ݅ሻǡ ʹǡ ܨ ሻ.
Now, we briefly describe how to produce all
possible legal sequences of length ݅ ͳ from all
previously created legal sequences of length ݅. Let
ܮ be the set of all extended mutation fragments at
column ݅, i.e., each element in ܮ corresponds to
some legal sequence of length ݅. Suppose that ܮ
x ࣦሺ4ǡ ݔǡ ܨሻ ൌ
݈
ቐ
݅
ߠǡ ߬ǡǡ א4Ǥ Ǥ ܨൌ ࣠൫ߠǡ ǡ ݔǡ ݅൯
ܨൌ ࣠൫߬ǡǡ ǡ ݔǡ ݅൯
x ࣬ሺ4ǡ ݔǡ ܨሻ ൌ
ݎ
ቐ
݅
ߠǡ ߬ǡǡ א4Ǥ Ǥ ܨൌ ࣠൫ߠǡ ǡ ݔǡ ݅൯
ܨൌ ࣠൫߬ǡǡ ǡ ݔǡ ݅൯
For example, suppose that 4 ൌ ൛߬ଵǡଷǡଶ ǡ ߠସǡହ ൟ ,
ݔൌ ܶ ܥܣܩܣand ࣠ሺ4ǡ ݔሻ ൌ ۦሺͳǡʹǡ ܣሻǡ ሺʹǡ͵ǡ ܩሻǡġ
ሺ͵ǡͳǡ ܶሻǡ ሺͶǡͷǡ ܩሻǡ ሺͷǡͶǡ ܶሻۧǤ For ܨൌ ሺʹǡ͵ǡ ܩሻ, we
have ࣦሺ4ǡ ݔǡ ܨሻ ൌ ͳ and ࣬ሺ4ǡ ݔǡ ܨሻ ൌ ͵.
For an integer ݅, let ܣ௫ ൌ ൛൫݅ ͳǡ ݐǡ ߠሺݔ௧ ሻ൯
݅ ͳ ൏ ݐ ݊ൟ and ܤ௫ ൌ ሼሺ݅ ͳǡ ݐǡ ݔ௧ ሻ ݅ ͳ ൏
ݐ ݊ሽ , which are sets of mutation fragments
covered by inversion and transposition operations
respectively acting on ݔwhose left end points are
all ݅ ͳ, where Ͳ ݅ ൏ ݊.
Definition 3. Let ܵ ൌ ܨۃଵ ǡ ܨଶ ǡ ǥ ǡ ܨ ۄbe an
agreed sequence of ݉ mutation fragments of ݔ,
where ݉ ݊, and 4 be a set of non-overlapping
mutation operations such that ܵ ٌ ࣠ሺ4ǡ ݔሻ .
Sequence ܵ is called a complete sequence if
࣬ሺ4ǡ ݔǡ ܨ ሻ ൌ ݉.
Observation 2. Suppose that ܵ ൌ ܨۃଵ ǡ ܨଶ ǡ ǥ ǡ ܨ ۄ
is a complete sequence, where ݉ ൏ ݊. Then ܵ ᇱ ൌ
ܨۃଵ ǡ ܨଶ ǡ ǥ ǡ ܨ ǡ ܨାଵ ۄis an agreed sequence if
ܨାଵ ܣ א
௫ ܯ௫ ሾͳǡ ݉ ͳǡ ݉ ͳሿ ܤ௫
ܯ௫ ሾʹǡ ݉ ͳǡ ݉ ͳሿ.
Definition 4. (Legal sequence) An agreed
sequence ܵ ൌ ܨۃଵ ǡ ܨଶ ǡ ǥ ǡ ܨ ۄof ݉ mutation
fragments of ݔis legal if κሺSሻ ൌ ݕଵǡ , where
݉ ݊.
Observation 3. Assume that ܵ ൌ ܨۃଵ ǡ ܨଶ ǡ ǥ ǡ ܨ ۄ
is a legal sequence. Then ܵԢ ൌ ܨۃଵ ǡ ܨଶ ǡ ǥ ǡ ܨିଵ ۄ
is also a legal sequence and κሺܨ ሻ ൌ ݕ , where
ͳ ൏ ݉ ݊.
The main idea of our algorithm is as follows.
First, the algorithm constructs all legal sequences
of length ͳ by examining all mutation fragments
in ܣ௫ ܯ௫ ሾͳǡͳǡͳሿ ܤ௫ ܯ௫ ሾʹǡͳǡͳሿ. A legal
sequence is then created if the mutation fragment
yields ݕଵ . Next, for each ͳ ݅ ൏ ݊ , the
algorithm produces all possible legal sequences of
58
The 32nd Workshop on Combinatorial Mathematics and Computation Theory
is already known. Below, we construct ܮାଵ by
examining all extended mutation fragments in ܮ .
For each extended mutation fragment ݁ ܮ א , we
identify all candidate mutation fragments at
column ݅ ͳ of the mutation tables ܯ௫ based
on the guiding information contained in ݁. Let
ܰ௫ ሺ݁ሻ be set of the candidate mutation fragments
of ݔat column ݅ ͳ. Then we compute ܰ௫ ሺ݁ሻ
as follows:
Case 1: ݁ is a continuing fragment. Let ܨ ൌ
ሺ݅ǡ ݆ǡ ߪሻ be the mutation fragment contained in ݁.
Then we consider three sub-cases to compute
ܰ௫ ሺ݁ሻ: (1) Suppose that ݁ is of type ܫ. Then
ܰ௫ ሺ݁ሻ ൌ ൛ሺ݅ ͳǡ ݆ െ ͳǡ ߠሺݔିଵ ሻሻൟǤ (2) Suppose
that ݁ is of type ܨ, i.e., ݁ ൌ ሺݏǡ ݀ǡ ͳǡ ܨ ሻ. If ݆ ൏
݊ ,
then
ܰ௫ ሺ݁ሻ ൌ ൛ሺ݅ ͳǡ ݆ ͳǡ ݔାଵ ሻǡ ሺ݅
ͳǡ ݏǡ ݔ௦ ሻൟ ; otherwise (i.e., ݆ ൌ ݊ ), ܰ௫ ሺ݁ሻ ൌ
ሼሺ݅ ͳǡ ݏǡ ݔ௦ ሻሽ. (3) Suppose that ݁ is of type ܵ,
i.e., ݁ ൌ ሺ݃ǡ ݀ǡ ʹǡ ܨ ሻ. Then ܰ௫ ሺ݁ሻ ൌ ൛ሺ݅ ͳǡ ݆
ͳǡ ݔାଵ ሻൟ.
Case 2: ݁ is an ending fragment. We set
ܰ௫ ሺ݁ሻ ൌ ܣ௫ ܯ௫ ሾͳǡ ݅ ͳǡ ݅ ͳሿ ܤ௫
ܯ௫ ሾʹǡ ݅ ͳǡ ݅ ͳሿ.
For any ܨାଵ ܰ א௫ ሺ݁ሻ with κሺܨାଵ ሻ ൌ ݕାଵ , an
extended mutation fragment ݁Ԣ is created to
contain ܨାଵ . As to the values of the guiding
information in ݁Ԣ, they can easily be obtained
from those in ݁ based on Observations 1 and 2.
Finally, we add ݁Ԣ into ܮାଵ .
for convenience. Based on Lemma 1, we have the
following corollary.
Corollary 1. Let ݁ଵ and ݁ଶ be two ending
fragments in ܮ . If ݀ሺ݁ଵ ሻ ൏ ݀ሺ݁ଶ ሻ, then we can
safely discard ݁ଶ from ܮ when computing the
mutation distance between ݔand ݕ.
Lemma 2. Let ݁ଵ and ݁ଶ be two continuing
fragments ݁ଵ and ݁ଶ of the same type ܵ in ܮ
and
݁ଶ ൌ
with
݁ଵ ൌ ൫݃ǡ ݀ଵ ǡ ʹǡ ሺ݅ǡ ݆ǡ ݔ ሻ൯
൫݃ǡ ݀ଶ ǡ ʹǡ ሺ݅ǡ ݆ǡ ݔ ሻ൯. If ݀ሺ݁ଵ ሻ ൏ ݀ሺ݁ଶ ሻ (i.e., ݀ଵ ൏
݀ଶ ), then we can safely remove ݁ଶ from ܮ
without affecting the computation of the mutation
distance between ݔand ݕ.
Proof. Since two continuing fragments ݁ଵ and ݁ଶ
of type ܵ contain the same mutation fragment
and guiding information ݃, the right end point of
the mutation operation covering ሺ݅ǡ ݆ǡ ݔ ሻ in ݁ଵ is
equal to that of the mutation operation covering
ሺ݅ǡ ݆ǡ ݔ ሻ in ݁ଶ . Denote by ݐthe above right end
point. Therefore, the candidate mutation fragments
from column ݅ to column ݐon the mutation
table ܯ௫ are exactly equivalent. Then we can
safely discard ݁ଶ from ܮ if ݀ଵ ൏ ݀ଶ according
to Corollary 1. Based on Corollary 1, we can also verify that
there does not exist two continuing fragments of
same type ܫor ܨin ܮ such that the values of
all their components, excluding the second
components (i.e., the number of used mutation
operations), are equal. Now, the details of our
algorithm for computing non-overlapping
inversion and transposition distance between two
strings is described in Algorithm 1.
Lemma 1. Let ܲଵ and ܲଶ be two complete
sequences of length ݉ of ݔwith κሺܲଵ ሻ ൌ κሺܲଶ ሻ.
Moreover, let ܵଵ be an agreed sequence of ݔ
with ܲଵ ٌ ܵଵ and ܵଶ be the resulting sequence
obtained by replacing ܲଵ with ܲଶ in ܵଵ . Then,
ܵଶ is also an agreed sequence of ݔand κሺܵଵ ሻ ൌ
κሺܵଶ ሻ.
Proof. Since ܵଵ and ܲଶ are agreed sequences of
ݔ, there exist sets of non-overlapping mutation
operations 4ଵ and 4ଶ such that ܵଵ ٌ ࣠ሺ4ଵ ǡ ݔሻ
and ܲଶ ٌ ࣠ሺ4ଶ ǡ ݔሻ . Moreover, ܲଵ ٌ ࣠ሺ4ଵ ǡ ݔሻ
since ܲଵ ٌ ܵଵ . We construct a set 4ଷ of
non-overlapping mutation operations from 4ଵ as
follows. First, let 4ଷ be the resulting set obtained
by removing all mutation operations in 4ଵ that
intersect with the range ሾͳǡ ݉ሿ . Note that the
ranges of these removed mutation operations are
completely contained in the range ሾͳǡ ݉ሿ because
ܲଵ is a complete sequence of length ݉. Next, we
insert those mutation operations in 4ଶ whose
ranges entirely lie in the range ሾͳǡ ݉ሿ into 4ଷ . It
can be verified that 4ଷ is a set of
non-overlapping mutation operations and as a
result, ܵଶ ٌ ࣠ሺ4ଷ ǡ ݔሻ and κሺܵଵ ሻ ൌ κሺܵଶ ሻ. Given an extended mutation fragment݁, we use
݀ሺ݁ሻ to indicate the value of its second component
(i.e., the number of the used mutation operations)
Algorithm
1:
Computing
non-overlapping
inversion
and
transposition distance between two
strings
Input: Two strings ݔand ݕof the
same length ݊.
Output: Mutation distance ݉݀ሺݔǡ ݕሻ.
1) Let ݅ ൌ Ͳ, ݀ ൌ Ͳ and ܮଵ ൌ ;
2) Construct ܮଵ by calling
EMF_Creationሺ݅ǡ ݀ሻ;
3) if ܮଵ ൌ then return f;
4) for ݅ ൌ ͳ to ݊ െ ͳ do
5)
ܮାଵ ൌ EMF_Extensionሺܮ ሻ;
6)
if ܮାଵ ൌ then return f;
7) end for
8) return ሼ݀ሺ݁ሻ ܮ א ݁ ሽ;
Procedure 1: EMF_Creationሺ݅ǡ ݀ሻ
Input: Column ݅ and current number
of mutation operations ݀.
59
The 32nd Workshop on Combinatorial Mathematics and Computation Theory
19)
case 3: ݁ is of type ܫ, let
݁ ൌ ቀ݃ǡ ݀ǡ ൫݅ǡ ݆ǡ ߠሺݔ ሻ൯ቁ.
20)
if ݃ Ͳ then
21)
if ߠ൫ݔିଵ ൯ ൌ ݕାଵ then
22)
Add ቀ݃ െ ͳǡ ݀ǡ ൫݅ ͳǡ ݆ െ
ͳǡ ߠሺݔିଵ ሻ൯ቁ of type ܫto ܮାଵ ;
23)
end if
24)
else
//݃ ൌ Ͳ
25)
Add ݁ to ܧ based on
Corollary 1;
26)
end if
27)
case 4: ݁ is of type ܸ.
28)
Add ݁ to ܧ based on
Corollary 1;
29) end for
30) if ܧ ് then
31)
݀ ൌ ሼ݀ሺ݁ሻ ܧ א ݁ ሽ;
32)
EMF_Creationሺ݅ǡ ݀ሻ;
33) end if
34) return ܮାଵ ;
Output: Extended mutation fragments
to be added to ܮାଵ when there is an
ending fragment at column ݅.
1) for each ൫݅ ͳǡ ݐǡ ߠሺݔ௧ ሻ൯ ܣ א௫ do
2)
if ߠሺݔ௧ ሻ ൌ ݕାଵ then
3)
Add ൫ ݐെ ݅ െ ͳǡ ݀ ͳǡ ሺ݅
ͳǡ ݐǡ ߠሺݔ௧ ሻሻ൯ of type ܫto ܮାଵ ;
4)
end if
5) end for
6) if ߠሺݔାଵ ሻ ൌ ݕାଵ then
7)
Add ending fragment ൫Ͳǡ ݀
ͳǡ ሺ݅ ͳǡ ݅ ͳǡ ߠሺݔାଵ ሻሻ൯ of type ܫto ܮାଵ
based on Corollary 1;
8) end if
9) for each ሺ݅ ͳǡ ݐǡ ݔ௧ ሻ ܤ א௫ do
10)
if ݔ௧ ൌ ݕାଵ then
11)
Add ൫݅ ͳǡ ݀ ͳǡͳǡ ሺ݅ ͳǡ ݐǡ ݔ௧ ሻ൯
of type ܨto ܮାଵ ;
12)
end if
13) end for
14) if ݔାଵ ൌ ݕାଵ then
15)
Add ending fragment ൫Ͳǡ ݀ǡ ሺ݅
ͳǡ ݅ ͳǡ ݔାଵ ሻ൯ of type ܸ to ܮାଵ based on
Corollary 1;
16) end if
Theorem 1. Algorithm 1 computes mutation
distance of two input strings in Oሺ݊ଷ ሻ time and
Oሺ݊ଶ ሻ space.
Proof. The correctness of Algorithm 1 can be
verified according to the discussion we mentioned
before. Below, we analyze the complexities of
Algorithm 1. For convenience, we use ȁܪȁ to
denote the number of extended mutation
fragments of type ܪin ܮ . By Corollary 1 and
Lemma 2, the numbers of extended mutation
fragments of type ܫ, ܨand ܵ in ܮ are ܱሺ݊ଶ ሻ.
Therefore, Procedure 2 can be done in ܱሺȁܫȁ
ȁܵȁ ȁܨȁሻ ൌ ܱሺ݊ଶ ሻ time. It is not hard to see that
Procedure 1 costs ܱሺ݊ሻ time in the worst case.
Moreover, Procedure 2 is called at most ܱሺ݊ሻ
times in Algorithm 1. As a result, the total time
complexity of Algorithm 1 is ܱሺ݊ଷ ሻ. In addition,
the space complexity of Algorithm 1 is ܱሺȁܫȁ
ȁܨȁ ȁܵȁሻ ൌ ܱሺ݊ଶ ሻ. Procedure 2: EMF_Extensionሺܮ ሻ
Input: ܮ .
Output:ܮାଵ .
1) Let ܧ be a set of ending
fragments at column ݅;
2) ܮାଵ m ; ܧ m ;
3) for each extended mutation
fragment ݁ in ܮ do
4)
case 1: ݁ is of type ܨ, let ݁ ൌ
൫ݏǡ ݀ǡ ͳǡ ሺ݅ǡ ݆ǡ ݔ ሻ൯.
5)
if ݆ ൏ ݊ and ݔାଵ ൌ ݕାଵ then
6)
Add ൫ݏǡ ݀ǡ ͳǡ ሺ݅ ͳǡ ݆ ͳǡ ݔାଵ ሻ൯
of type ܨto ܮାଵ ;
7)
end if
8)
if ݔ௦ ൌ ݕାଵ then
9)
Add ൫݆ െ ݅ െ ͳǡ ݀ǡ ʹǡ ሺ݅
ͳǡ ݏǡ ݔ௦ ሻ൯ of type ܵ to ܮାଵ , based on
Lemma 2;
10)
end if
11)
case 2: ݁ is of type ܵ, let ݁ ൌ
൫݃ǡ ݀ǡ ʹǡ ሺ݅ǡ ݆ǡ ݔ ሻ൯.
12)
if ݃ Ͳ then
13)
if ݔାଵ ൌ ݕାଵ then
14)
Add ൫݃ െ ͳǡ ݀ǡ ʹǡ ሺ݅ ͳǡ ݆
ͳǡ ݔାଵ ሻ൯ of type ܵ to ܮାଵ based on
Lemma 2;
15)
end if
16)
else
// ݃ ൌ Ͳ
17)
Add ݁ to ܧ based on
Corollary 1;
18)
end if
4
Conclusions
In this work, we studied the computation of
non-overlapping inversion and transposition
distance between two strings, which can be useful
applications especially in computational biology
(e.g., evolutionary tree reconstruction of biological
sequences). As a result, we presented an Oሺ݊ଷ ሻ
time and Oሺ݊ଶ ሻ space algorithm to compute this
mutation distance. Note that the mutation
operations we considered allow both inversion and
transposition and the lengths of two substrings
exchanged by a transposition are not necessarily
identical. It is worth mentioning that the algorithm
we proposed in this study can be used to solve the
approximate string matching problem under
60
The 32nd Workshop on Combinatorial Mathematics and Computation Theory
non-overlapping inversion and/or transposition
distance.
References
[1]
A. Amir, T. Hartman, O. Kapah, A. Levy, E.
Porat, On the cost of interchange rearrangement
in strings, SIAM Journal On Computing 39
(2009) 1444–1461
[2] D. Cantone, S. Cristofaro, S. Faro, Efficient
string-matching allowing for non-overlapping
inversions, Theoretical Computer Science 483
(2013) 85–95
[3] D. Cantone, S. Faro, E. Giaquinta,
Approximate string matching allowing for
inversions and translocations, Proceedings of
the Prague Stringology Conference 2010, pp.
37–51
[4] D.-J. Cho, Y.-S. Han, H. Kim, Alignment with
non-overlapping inversions on two strings, in:
S.P. Pal, K. Sadakane (eds.) WALCOM 2014,
LNCS , vol. 8344, Springer, Heidelberg, 2014,
pp. 261–272.
[5] D.-J. Cho, Y.-S. Han, H. Kim, Alignment with
non-overlapping inversions and translocations
on two strings, Theoretical Computer Science,
in press (2014)
[6] S. Grabowski, S. Faro, E. Giaquinta, String
matching with inversions and translocations in
linear average time (most of the time),
Information Processing Letters 111 (2011)
516–520
[7] S.-Y. Huang, C.-H. Yang, T.T. Ta, C.L. Lu,
Approximate string matching problem under
non-overlapping inversion distance, in:
Workshop on Algorithms and Computation
Theory in conjunction with 2014 International
Computer Symposium, 2014, pp. 38–46
[8] Y.-L. Huang, C.L. Lu, Sorting by reversals,
generalized transpositions, and translocations
using permutation groups. Journal of
Computational Biology 17 (2010) 685–705
[9] O. Kapah, G.M. Landau, A. Levy, N. Oz:
Interchange rearrangement: the element-cost
model, Theoretical Computer Science, 410
(2009) 4315–4326
[10] F.T. Zohora, M.S. Rahman, Application of
consensus string matching in the diagnosis of
allelic heterogeneity, in: Basu, M., Pan, Y.,
Wang, J. (eds.) ISBRA 2014. LNBI 8492, vol.
8344, Springer, Switzerland, 2014, pp. 163–175
61