AP Biology Corn Genetics and Chi
advertisement

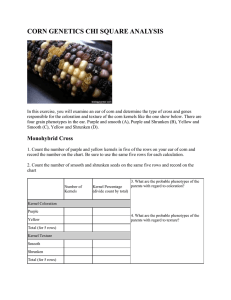
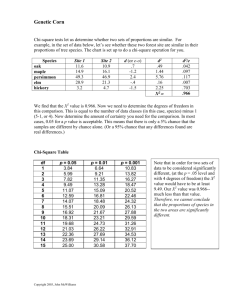
AP Biology Corn Genetics and Chi-Square Analysis In this exercise, you will examine an ear of corn and determine the type of cross and genes responsible for the coloration and texture of the corn kernels like the one show below. There are four grain phenotypes in the ear. Purple and smooth (A), Yellow and Smooth (B), Yellow and Shrunken (C), Purple and Shrunken (D). Monohybrid Cross (Corn 8910) 1. Count the number of purple and yellow kernels in five of the rows on your ear of corn and record the number on the chart. Be sure to use the same five rows for each calculation. 2. Count the number of smooth and shrunken seeds on the same five rows and record on the chart. Number of Kernels Kernel Percentage (divide count by total) Kernel Coloration Purple Yellow Total (for 5 rows) Kernel Texture Smooth Shrunken Total (for 5 rows) 3. What are the probable phenotypes of the parents with regard to coloration? 4. What are the probable phenotypes of the parents with regard to texture? Monohybrid Cross (Corn 8900) 5. Count the number of purple and yellow kernels in five of the rows on your ear of corn and record the number on the chart. Be sure to use the same five rows for each calculation. 6. Count the number of smooth and shrunken seeds on the same five rows and record on the chart. Number of Kernels Kernel Percentage (divide count by total) Kernel Coloration Purple Yellow Total (for 5 rows) Kernel Texture Smooth Shrunken Total (for 5 rows) 7. What are the probable phenotypes of the parents with regard to coloration? 8. What are the probable phenotypes of the parents with regard to texture? Dihybrid Cross (Corn 8915) 9. We will now consider a dihybrid cross, which is a combination of the two monohybrids. Your ear of corn may be a result of a cross between plants that were both heterozygous (PpSs x PpSs). Create a punnett square or use a mathematical system to determine the phenotype ratio. Record what you would expect to get from this cross in the chart below. 10. Purple & smooth ____ Purple & shrunken ______ Yellow & smooth _____ Yellow & shrunken ______ 11. Now count the number of each in your five rows on the ear of corn. Number Counted Purple & smooth Purple & shrunken Yellow & smooth Yellow & shrunken TOTAL Percentage: Number counted / total x 100 12. Did you obtain a 9:3:3:1 ratio? To determine if the deviations from your observed data are due to chance alone or if the data is significantly different, you need to use a chi square test. The table below will help you make the calculations. Expected Number Purple & smooth Total x 9/16 = Purple & shrunken Total x 3/16 = Yellow & smooth Total x 3/16 = Yellow & shrunken Total x 1/16 = Observed Number ÷ expected CHI SQUARE VALUE ========> (add the numbers from the rows above) 13. Now determine if your chi square value is a good fit with your data. Your degrees of freedom (df) is the number of possible phenotypes minus 1. In your case, 4 - 1 = 3. Find the number in that row that is closest to your chi square value. Circle that number. 14. Explain what it means to have a "good fit" or a "poor fit". Does you chi square analysis of real corn data support the hypothesis that the parental generation was PpSs x PpSs? Dihybrid Cross (Corn 8907) 15. We will now consider a dihybrid cross, which is a combination of the two monohybrids. Your ear of corn may be a result of a cross between plants that were PpSs x ppss). Create a punnett square or use a mathematical system to determine the phenotype ratio. Record what you would expect to get from this cross in the chart below. This is a dihybrid backcross (or test cross) example. 16. Purple & smooth ____ Purple & shrunken ______ Yellow & smooth _____ Yellow & shrunken ______ 17. Now count the number of each in your five rows on the ear of corn. Number Counted Purple & smooth Purple & shrunken Yellow & smooth Yellow & shrunken TOTAL Percentage: Number counted / total x 100 18. What ratio did you expect to obtain? Did you obtain this expected ratio? To determine if the deviations from your observed data are due to chance alone or if the data is significantly different, you need to use a chi square test. The table below will help you make the calculations. Expected Number Purple & smooth Total x _____ = Purple & shrunken Total x _____ = Yellow & smooth Total x _____= Yellow & shrunken Total x _____ = Observed Number ÷ expected CHI SQUARE VALUE ========> (add the numbers from the rows above) 19. Now determine if your chi square value is a good fit with your data. Your degrees of freedom (df) is the number of possible phenotypes minus 1. In your case, 4 - 1 = 3. Find the number in that row that is closest to your chi square value. Circle that number. 20. Explain what it means to have a "good fit" or a "poor fit". Does you chi square analysis of real corn data support the hypothesis that the parental generation was PpSs x ppss? Mystery Corn (Corn 8920) This ear of corn is an F2 level corn cross result showing the segregation of a dominant aleurone (purple) color gene (Yellow is dominant) and a dominant color-inhibiting gene. This is an example of epistasis. 21. These ears of corn may be a result of a cross between plants that were PyCn x PyCn. (P-purple, yyellow, C-colored, n-no color) Create a punnett square or use a mathematical system to determine the phenotype ratio. Record what you would expect to get from this cross in the chart below. This is a dihybrid backcross (or test cross) example. 22. Purple ____ Yellow _____ 23. Now count the number of each in your five rows on the ear of corn. Number Counted Purple Yellow TOTAL Percentage: Number counted / total x 100 24. What ratio did you expect to obtain? Did you obtain this expected ratio? To determine if the deviations from your observed data are due to chance alone or if the data is significantly different, you need to use a chi square test. The table below will help you make the calculations. Expected Number Purple Total x _____ = Yellow Total x _____= Observed Number CHI SQUARE VALUE ========> (add the numbers from the rows above) 25. Now determine if your chi square value is a good fit with your data. Your degrees of freedom (df) is the number of possible phenotypes minus 1. In your case, 2 - 1 = 1. Find the number in that row that is closest to your chi square value. Circle that number. 26. Explain what it means to have a "good fit" or a "poor fit". Does you chi square analysis of real corn data support the hypothesis that the parental generation was PyCn x PyCn? (Modified from Biology.com “Corn Genetics and Chi-square Analysis”: http://www.biologycorner.com/worksheets/corn_chi.html) ÷ expected