Lecture 19 Membrane Protein - The Center for Molecular and
advertisement

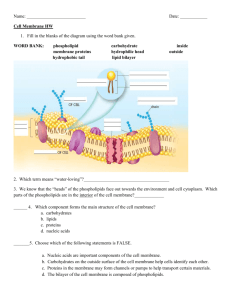
Lecture 19 Membrane Protein James Chou BCMP201 Spring 2008 Lecture outline Properties of cellular membrane, lipid bilayer, detergent micelles Membrane protein topologies Folding of proteins into membrane assisted by protein translocon Signaling across membrane: example of G protein coupled receptor Membrane protein is one of the frontiers in structural biology Membrane proteins account for 25-35% of genome in living organisms. There are over 17,000 structures of water-soluble proteins, but only ~150 unique structures of membrane proteins. http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html Many membrane-embedded receptors, transporters, and ion channels are important therapeutic targets. Properties of cellular membrane The Fluid Mosaic Model of plasma membrane, Singer & Nicholson, 1972 http://www.molecularexpressions.com/cells/plasmamembrane/plasmamembrane.html Proteins vs Lipid Protein Lipid Plasma Membrane 50% 50% Axon 20% 80% Mitochondria Membrane 75% 25% Assembly of lipid bilayer Common cartoon drawing of lipid bilayer Membrane lipid components Sterol Cholesterol Weight percent of membrane components in several types of membranes Alberts et al., Molecular Biology of the Cell 1994 Permeability of lipid bilayer Dynamic properties of lipid bilayer Common cartoon drawing of lipid bilayer Real lipid bilayer is dynamic MD simulation from Dr. Scott Feller, Wabash College Structure of a fluid DOPC bilayer Time averaged distributions of the principal structural groups of the lipid. Structure determined by joint refinement of X-ray and neutron diffraction data (Wiener and White, Biophys J, 1992). 3D viewing of the simulated DOPC bilayer MD simulation from Dr. Scott Feller, Wabash College Other membrane properties Rate of lateral diffusion of lipid molecules in E. coli membrane is 10-8 10-7 cm2s-1. Rate of flip-flop of lipids (from one half of a bilayer to the other) is very slow, e.g., time for movement of 50% of lipids from one monolayer to the other is hours to days. Lipid flip-flop in real membranes is catalyzed by enzymes such as flippases. Various lipids, detergents, metals, and proteins can induce membrane curvature Extract membrane proteins from cell membranes using detergents Fractionation of membranes using centrifuge Detergent micelle formation Property of detergent micelle C free = conc. of detergent not in micelle Cmic = conc. of detergent micelle N = aggregation number Ctotal = NCmic + C free Critical micelle conc. (cmc) is Ctotal at which C free = NCmic Lipid/detergent bicelles With detergent Without detergent Various detergent micelles and lipid/detergent bicelles Micelle / Bicelle q CMC Mass [lipid]/[detergt] (mM) (kD) - 25 23.0 DPC - 1 19.0 DHPC - 14 12.3 CHAPSO - 8 6.0 - 2 50.0 - 0.05 64.6 DMPC/DHPC 0.15 10 21.7 POPC/DHPC 0.15 10 23.4 POPC/DHPC 0.30 8 31.8 POPC/DHPC 0.50 7 60.2 -Octylglucoside -decylmaltoside 16:0 Lyso PG (LPPG) Vinogradova et al., Biochemistry 1998; Chou et al., JBNMR 2004 Membrane protein topology History of membrane protein structure determination 1984 Photosynthetic reaction centre, Deisenhofer et al, JMB 1984 1990 Bacteriorhodopsin, Henderson et al, JMB 1990 1992 Porin (beta-barrel), Weiss & Schulz, JMB 1992 1998 K+ channel, Doyle et al, Science 1998 2000 Rhodopsin, Palczewski et al, Nature 2000 Example of a helical membrane protein: the mitochondrial ATP/ADP carrier Pebay-Peyroula et al, Nature 2003 Example of a beta barrel membrane protein: the outer membrane protein G (OmpG) Subbarao and van den Berg, JMB 2006 Some definitions of membrane protein types Type I Type II Type III Hydropathy plot http://www.vivo.colostate.edu/molkit/hydropathy/index.html Hydrophobicity scale of amino acids Amino Acid Ala Arg+ Asn AspAsp0 Cys Gln GluGlu0 Gly G(interface) (kcal/mol) 0.17+0.06 0.81+0.11 0.42+0.06 1.23+0.07 -0.07+0.11 -0.24+0.06 0.58+0.08 2.02+0.11 -0.01+0.15 0.01+0.05 G(octanol) (kcal/mol) 0.50+0.12 1.81+0.13 0.85+0.12 3.64+0.17 0.43+0.13 -0.02+0.13 0.77+0.12 3.63+0.18 0.11+0.12 1.15+0.11 His+ His0 Ile Leu Lys+ Met Phe Pro Ser Thr Trp Tyr Val White and Wimley, 1999 0.96+0.12 0.17+0.06 -0.31+0.06 -0.56+0.04 0.99+0.11 -0.23+0.06 -1.13+0.05 0.45+0.12 0.13+0.08 0.14+0.06 -1.85+0.06 -0.94+0.06 0.07+0.05 2.33+0.11 0.11+0.06 -1.12+0.11 -1.25+0.11 2.80+0.11 -0.67+0.11 -1.71+0.11 0.14+0.11 0.46+0.11 0.25+0.11 -2.09+0.11 -0.71+0.11 -0.46+0.11 Program: Membrane Protein Explor Transmembrane helix prediction Performance comparison of various TMH predictors a Predictor VTMH VP N-score C-score free-R THUMBU[16] b 85.5% 47.1% 6.9 6.7 0.58±0.04 SOSUI[11] c 89.1% 57.1% 5.0 5.0 0.44±0.04 DAS-TMfilter[20] d 90.7% 64.3% 6.5 5.5 0.58±0.03 TOP-PRED[1] e 92.6% 60.0% 4.5 4.6 0.45±0.02 TMHMM[6] f 91.0% 65.7% 4.5 4.5 0.44±0.02 Phobius[7] g 91.8% 71.4% 4.6 4.4 0.44±0.04 MemBrain h 97.9% 87.1% 3.2 3.1 0.35±0.02 a The testing dataset consists of 378 TMH segments from 70 proteins (see Supplementary Table S2). b http://sparks.informatics.iupui.edu/Softwares-Services_files/thumbup.htm [16]. c http://bp.nuap.nagoya-u.ac.jp/sosui/ [11]. d http://mendel.imp.ac.at/sat/DAS/DAS.html [20]. e http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html [1]. f http://www.cbs.dtu.dk/services/TMHMM/ [6]. g http://phobius.cgb.ki.se/ [7]. h http://chou.med.harvard.edu/bioinf/MemBrain/. The Positive Inside Rule cytoplasm + + + + - + + von Heijne, G. EMBO J 1986. Pseudosymmetry Many helical membrane proteins (~60%) contain homologous domains with opposite membrane orientation. Example: 3D viewing of Aquaporin 1 (6 TMs), PDB code : 1J4N. Sui et al, Nature 2001. How do membrane proteins fold into lipid bilayer? ? Image from University of Oxford Protein translocation across the plasma membrane (co-translational translocation) (Signal Recognition Particle) ER membrane Rapoport, T. A., Nature 2007 Protein translocation across the plasma membrane (post-translational translocation in bacteria) Cytosolic ATPase Protein translocation into the plasma membrane ribosome 3D viewing of protein translocon from the archaeon Methanococcus jannaschii, Sec YEβ van den Berg et al, Nature 2004. PDB code : 1RHZ EM structure of protein translocon in complex with ribosome in E. coli Mitra et al., Nature 2005 The MD simulation system Total 106,679 atoms 251 POPC lipids 21520 H2O 5 Na+ 21 ClProtein SecYEβ 111 X 109 X 106.5 A3 Gumbart and Schulten, Biophys J. 2006 MD simulation of protein translocation across the protein translocon Force applied in Steered MD (~10 ns simulation) MD simulation of the lateral exit of peptide from protein translocation MD simulation of the lateral exit of peptide from protein translocation (~1 us) Gumbart and Schulten, Biochemistry 2007 Membrane receptors Two types of membrane receptors which transduce signal across the membrane ligand ligand Y Y Y Y phosphorylation activation of other proteins Example: TCR/CD3 signaling α β ε γ Y Y α β ε δ ζζ ε γ Y Y Y Y Y phosphorylation ε δ ζζ Y Y Y Y Y Example: G protein coupled receptors Seven transmembrane helices Largest family of membrane receptors 50% of medicinal targets Difficult to express and crystallize Coupling to different receptor isoforms lead to different G protein pathways and different biological effects General themes in heterotrimeric G protein pathways Rockman H.A. et al (2002) Nature 415:206 Rhodopsin - the visual pigment of rod cells Palczewski et al, Science 2000 Differences between the ground and photoactivated states disappeared changed Salom et al, PNAS 2006 3D viewing of Rhodopsin PDB code : 1F88 β2-adrenergic GPCR adrenalin Crystallization of β2-adrenergic GPCR V. Cherezov et al., Science 318, 1258 -1265 (2007) Ligand-binding characterization and comparison to rhodopsin