Lecture #5 – Relaxation What do we mean by
advertisement

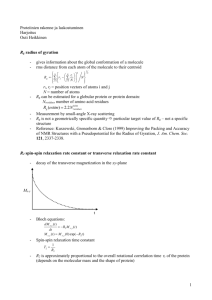
Lecture #5 – Relaxation 2/16/16 Clubb Topics Covered • Relaxation Time Constants (T1 and T2) • Mechanisms of relaxation (dipole-dipole) • Relaxation protein/nucleic acid dynamics Why do we care about NMR relaxation? 1) Various relaxation rates report on molecular dynamics 2) Dipolar relaxation is the basis of the NOE: crucial to structure determination by NMR Reading: Claridge p 25-38 What do we mean by “relaxation”? Typical one dimensional (1D) experiment 90o (FID) Delay to enable system to relax back to equilibrium 90o Return to equilibrium via relaxation processes Mz=Mo equilibrium Mz=0 Mz=Mo equilibrium 1 Main Relaxation Parameters Measured in NMR T1: Longitudinal (also called spin-lattice) relaxation. It is a measure of rate of return of the magnetization to the Z-axis. Importance: 1) Dynamics 2) Determines delay times between subsequent NMR experiments 90o 90o (FID) (FID) ~900ms ~100ms Total recycle time ~1 s T2: Transverse (also called spin-spin) relaxation. It is a measure of The rate of decay of magnetization in the x-y plane. Importance: 1) Dynamics 2) Determines line-width of peaks 3) Can limit types of expts that can be performed Linewidth 1/ ( x T2*) FID Note: formula describes reappearance of Mz when it starts from X-Y plate relaxation Mz Mz=Mo Mz=0 Mz=Mo 2 •During T1 relaxation the spins lose energy to the lattice (surroundings) as a result of spins “flipping” from the high energy (beta) state to lower energy (alpha) state •Spins “flip” during relaxation process as a result of fluctuating FID magnetic fields that generate RF at the appropriate frequency T1 relaxation Mz=Mo Mz=Mo Mz = 0 Physical origin of oscillating magnetic fields that cause relaxation: Dipole-dipole (dipolar) interactions Each atom produces its own magnetic field Tumbling of protein Tumbling of protein causes magnetic field from proximal atoms to vary. 3 Other sources of fluctuating magnetic fields that cause relaxation (not as important as dipolar relaxation in macromolecules) Chemical shift anisotropy (CSA). Negligible for protons, but important for nuclei with large chemical shift ranges such as carbon-13. Quadrupolar (negligible in proteins). Atoms with I > ½ have non-spherical charge. Interaction of resultant quadrupole moment with electric field gradients can cause relaxation. Scalar relaxation (negligible in proteins): caused by time dependent fluctuations in coupling constants Paramagnetic (negligible for most proteins): Relaxation caused by very large magnetic moment of unpaired electrons. Similar to nuclear dipole-dipole interaction. Not significant in most proteins because they do not contain unpaired electrons. T1 importance: Choose recycle times in multi-scan experiments to be ~5x T1 value of 1H atom Total Recycle time FID t Scan #1 t t #2 #3 #4 Delay between scans to enable relaxation (t) Summed FID •Choose total recycle time to be ~5x T1 value of 1H atom •1H T1 times in proteins are typically ~200ms FT NMR Spectrum 4 Recall: Recycle time = 5 * T1 Describes return of magnetization to Z-axis when it starts from the XY plane At recycle time = 5 * T1 e(-t/T1) = 0.0067 Mz ~ M0 When Mz = Mo system is at equilibrium Inversion recovery experiment is used to quantitatively measure T1 180o 90o Perform a series of 1D experiments using different times for 5 Inversion recovery spectra at various values of the delay when Mt = 0 Note: formula describes reappearance of Mz when it starts from –Z axis Review: p Pulse of duration p and strength B1 Pulses of different flip angles dM(t)/dt = M(t) X B1 6 Mz = -Mo at very long values Mz = Mo complete recovery to Mo occurs FT Detector At shorter values Mz does not fully recover to Mo. Can give rise to negative signal Short values FT Mz = -Mo Mz > -Mo Very long values Detector FT 7 Recall: Sign and phase of signal depends upon where magnetization starts at the beginning of detection. Detector T2 relaxation constant (spin-spin or transverse): Time constant describing rate of decay in the transverse (XY) plane. Larger T2 values slower decay of magnetization 8 Rate of dephasing characterized by the time constant T2 (transverse relaxation time or spin-spin relaxation time) * Linewidth at half-maximal intensity 1/ ( x T2*) Intrinsic time constant for decay (transverse relaxation time, also called spin-spin relaxation time) Signal decay caused by field inhomogeneity Spin-Echo experiments to selectively measure intrinsic T2 value of a nucleus. Experiment eliminates contribution from field inhomogeneity Dephasing from field inhomogeneity Field inhomogeneity effects refocused. However intrinsic dephasing caused by T2 is not. Therefore the magnitude of the vector has decreased. (note in relation to a simple one-pulse experiment the spectrum would be of opposite sign. This can be corrected by properly phasing the spectrum 9 Behavior of magnetization during the experiment In real experiment you begin detection of signal here. The FT of the resultant FID is a regular 1D spectrum. However the amplitude of the peaks are reduced as a result of T2 relaxation Behavior of signal during experiment Begin detection here n=5 10 FT Intensity α e –((time)/T2)) n=2 n=4 n=8 time time = n x (2+180 pulse) n=16 Relaxation and relationship to motion ( c ) molecular correlation time: time required for molecule (protein) to rotate through one radian a : viscosity a: radius k: Boltzman constant T: temperature 11 12 Overview: Relaxation Motion T2 (J()) and T1 (J()) Equations exist that relate experimentally measured relaxation parameters to J(). Relaxation Parameters (measured as T1 and T2) Spectral density function(J()) Fourier transform of autocorrelation function. Describes frequencies of motions that can promote relaxation Autocorrelation function G() Describes relationship between protein dynamics and fluctuating magnetic fields. Assumes motional models (e.g. molecule tumbles as sphere or as an elipsoid) fluctuating magnetic fields cause relaxation Motions within macromolecule create fluctuating fields Spectral Density Function (J()): Power available from the lattice (molecular motions) to bring about relaxation via transition probablities. It is a function of frequency (). All measurable relaxation properties of a protein (T1, T2, NOE) can be expressed in terms of the spectral density function. J(I s) = J (~0) • J() depends on how fast the macromolecule tumbles in solution. It therefore depends on the size and shape of the macromolecule, and the temperature and viscosity of the solution. J(I s) On 500 MHz instrument J(I s) = J (1GHz) For spherically shaped macromolecule ( isotropically tumbling) J() = c/(1+2c2) c ~100 MHz c = 10 ns 13 Example: T1 dependence on J() Relaxation depends upon: c, r, and o T1 ~ Low frequency motions Mid-range frequency motions (on a 500MHz spectrometer = 2 * 500 MHz) High frequency motions (on a 500MHz spectrometer = 2 * 1,000 MHz) Representative Spectral Density curves for three molecules of different sizes (assumes identical larmor frequencies) Big molecules Small molecules 14 Later…Nitrogen-15 Relaxation time constants • To gain insights into protein dynamics the T1 and T2 time constants of backbone nitrogen-15 nuclei are typically measured. • This nucleus is primarily relaxed by its directly bonded hydrogen-atom • The T1 and T2 values report upon its effective correlation time (eff), which is a combination of the overall molecular tumbling of the protein and internal motions it experiences. H Bond vector eff -1 = c-1 + e-1 N Protein 15