Growth and enumeration of viable cells and unknown two
advertisement

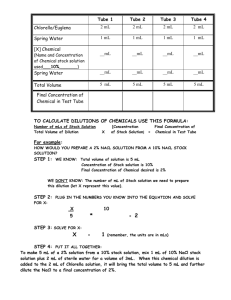
MMBB255 Week 5 Background 1 Growth and enumeration of viable cells and unknown two I. Objective The goal of this experiment is examine the kinetics of bacterial growth and to learn how to determine the number of cells in a broth culture using a spectrophotometer and to correlate this value with the number of colony forming units that result from viable cells. You will start unknown number two and follow up on several of the previous experiments. II. Background The growth rate of microbial cells is highly variable doubling time: the time it takes for a and depends on factors that include the type of organism, population of cells to double in number. the pH of the medium, the temperature, and nutritional factors. For example, Escherichia coli, an organism found in the intestinal tract, grows with a doubling time of about 30 min at 37°C in rich medium at pH 7 containing a carbon source such as glucose. If incubated at 30°C, the same organism in the same medium would grow with a doubling time >40 min. In contrast, a microbe isolated from the soil might grow best at 30°C and have a doubling time of 4-8 hours. In order to determine the doubling time of an organism growth curves are required. A growth curve is generated when you monitor the cell number over time. Cell numbers are generally on the y-axis and are in base10 log while time is on the x-axis in normal form (this is called semilog graphing; one axis is in log while the other is arithmetic). A typical growth curve will have four phases: the lag phase (where the cells are adapting to the growing conditions), the exponential phase (where the cells have unrestricted growth), the stationary phase (where the cells have run out of food or toxic by-products have accumulated), and the death phase (death and cell lysis). The doubling time is determined from the exponential phase of growth. In your professional capacity, whether it be medicine, environmental science, food microbiology, or research, it will be important that you know how to quantify microbes in a sample. For example, if your job involves advising the public of water quality standards, you must be able to report the numbers of microorganisms in a water sample and determine if the numbers are within a safe limit for human consumption. The following topics summarize the most common quantification procedures: A. Quantifying total cell number using a spectrophotometer We will use a The detector in the spectrophotometer this week to spectrophotometer receives all of estimate cell numbers based on the the light because water does not principle that cells absorb light. The absorb. sample to be measured is placed %T = 100 or A = 0.0 between a light source and a Water Light Source detector. The detector measures The detector receives less light light. The spectrophotometers that because the microbial cells you will use have two scales: one absorb some of the light. The scale indicates the amount of light amount of light absorbed absorbed by the sample called depends on the cell density. absorbance and the other is used to Cells measure the amount of light transmitted (passes through) by a sample called percent transmittance. A sample, such as air or water, which does not absorb light will have an absorbance = 0. (Absorbance = Abs or Awavelength and in older literature is called optical density or OD). Because air and water do not absorb light, all of the light will pass through to the detector. Hence, the % transmittance (%T) of air is 100%. We will measure absorbance of cells at 600 nm. We use this wavelength because absorbance of light by other molecules in the cell, such as flavins and carotenoids, is minimal at this position. Equations that relate OD to %T are provided in the box; Beer’s law gives a direct linear relationship of Abs to concentration and the abbreviation definitions are below: I0 = light before sample I = light that passes through the sample a = molar absorptivity in M-1cm-1 b = light pathlength of the sample in cm c = concentration in M MMBB255 Week 5 Background 2 This method can be used to estimate the total number of cells in a sample, which includes living cells, dead cells, and noncellular material that absorbs/scatters the light. To correlate the Abs (absorbance) with numbers of viable cells (living cells capable of cell division) we will be doing plate counts. This correlation is be done by removing an aliquot of cells from the growing culture, taking a spctrophotmetric reading, then diluting the cells, and then plating the dilutions (either by spread plate or pour plate, methods). Each viable cell will be able to grow in the agar medium and will produce a colony. These colonies, now called a colony forming unit or CFU, are counted and then calculated to get the cell concentrations. B. Quantification of cells via direct and electronic methods There are several methods for direct counting of total cell number. One of the most common is to count them by eye under the microscope using a Petroff-Hauser chamber. A Petroff-Hauser chamber has an exact grid etched into a glass slide. Cells are placed on the grid, covered with an absolutely flat cover slip, and counted within the gridded spaces. The grid with the cover slip gives a known volume which when combined with the number of cells counted in the grid will give you the concentration (cells/ml) of total cell number. Again, this is a total cell count that includes both viable (live) and non-viable (dead) cells. An electronic method uses the fact that cells have a different conductivity than an aqueous solution. Two chambers are connected by a small hole just big enough to let a cell through. A known volume is pumped through the hole and when a cell enters the hole the conductance changes and is counted. C. Quantification of cells by viable plating techniques The most accurate method for determining the number of viable (living cells capable of replication) cells is by plating aliquots of cell dilutions. There are various ways of plating the aliquots; the spread plate method, the pour plate method, and calibrated loop method are some examples. After a period of time (usually 24-48 hrs), the number of colonies (called colony forming units or cfu) on each plate can be counted. Only statistically valid plates, those that have between 25 and 250 colonies per plate, are counted (it used to be 30-300). This method is based on the assumption that each colony arose from a single cell. Hence, by counting the number of colonies and factoring in the various dilutions that were performed, you can estimate (extrapolate) the number of viable cells in the starting material (see the equations to the right). Dilutions and extrapolations generated from dilution series are particularly important in medicine, immunology, food microbiology, and ecology Here is an example of a dilution series and the pour plate method of plating to determine the number of viable cells in a sample: 1. A water sample (tube A) from a contaminated aquifer has an Abs600= 0.17 (this will look a little cloudy or turbid). You do a dilution series: You add 1 ml of the sample to 99 ml of diluen You add 1 ml of dilution 1 to 99 ml of diluent You add 1 ml of dilution 2 to 9 ml of diluent You add 1 ml of dilution 3 to 9 ml of diluent dilution 1 (tube B) a 1:100 dilution for a 10-2 total dilution dilution 2 (tube C) a 1:100 dilution for a 10-4 total dilution dilution 3 (tube D) a 1:10 dilution for a 10-5 total dilution dilution 4 (tube E) a 1:10 dilution for a 10-6 total dilution 2. 1 ml of dilutions 2, 3, and 4 were added aseptically to empty, sterile petri plates and tryptic soy agar (TSA), at 55°C, was immediately aseptically poured into each petri dish, mixed in a figure eight, and allowed to solidify. The plated sample was inverted and incubated at 37°C. The next day, colonies were counted. Your counts were: plate 1 (from tube C) plate 2 (from tube D) plate 3 (from tube E) = >>1000 (too numerous to count TNTC) = 450 (TNTC-not statistically valid) = 56 These are good results because plate 3 contains a statistically valid number of colonies to count. Statistically valid counts are between 25 and 250 colonies per plate. 3. Extrapolating, calculating, the original sample's viable cell number is next. Using the number of colonies from plate 3, you plug it into the equation here along with the corresponding total dilution and volume of sample added to the plate (see the work to the right). MMBB255 Week 5 Background 3 Tuesday’s Procedures: A. Practice streak: examining – Finished. 1. Remove your streak plate from the bucket. You should see three different colony morphologies. 2. Examine the colony morphology. Record your colony observations (week 1 has the criteria). Decide if you need to improve your streaking technique and how you would. Colony type 1 Colony type 2 Colony type 3 Colony description: morphology, color, texture, size etc. B. Technique: Dilutions and the spread plate method – Work in Pairs. Background: Another way to obtain isolated colonies is to spread a small amount of liquid containing cells on a plate. This is the spread-plate technique. Before spreading, you will need to make a few serial dilutions of your cell culture as it contains millions of bacteria per ml. You must understand how to perform a serial dilution and how to quantify using this technique. There are practice problems that are highly recommended for you to work out. 1. Make serial dilutions of the cell mixtures. Label the dilution tubes you prepared last Tuesday with numbers 1 through 8. Although you added 10 ml peptone water (the diluent) to each tube, some liquid is lost during the autoclaving process. As a result, each tube contains about 9 ml of peptone water, which is ideal for the dilutions we want to make. The diagram to the right is an example to help you visualize the procedure. a. Mix on the vortexer and remove 1 ml of sample that you used to practice streak last week (a mixture of three organisms). Add it to the 9 ml of peptone water in tube #1. Use aseptic technique. Your lab instructor will demonstrate the transfer technique. Remember to dispose the used pipette tips into the small biohazard bags. b. Mix tube 1 on the vortexer (this is crucial). This is a 1:10 dilution (also written as 1/10 or 10-1) for a total dilution of 10-1 c. Take a new sterile tip and remove 1 ml from tube 1 and add it to 9 ml (1:10) of peptone water in tube #2. This is a 1:10 dilution for a total dilution of 10-2 (relative to the starting material). MMBB255 Week 5 Background 4 d. Continue as follows. Using a new tip for each tube and mixing each tube before aseptically transferring; until the sample has been transferred to the last tube (as below): Add 1 ml from tube 2 to the 9 ml of peptone water in tube #3. Add 1 ml from tube 3 to the 9 ml of peptone water in tube #4. Add 1 ml from tube 4 to the 9 ml of peptone water in tube #5. Add 1 ml from tube 5 to the 9 ml of peptone water in tube #6. Add 1 ml from tube 6 to the 9 ml of peptone water in tube #7. Add 1 ml from tube 7 to the 9 ml of peptone water in tube #8. 1:10 dilution for a 10-3 total dilution 1:10 dilution for a 10-4 total dilution 1:10 dilution for a 10-5 total dilution 1:10 dilution for a 10-6 total dilution 1:10 dilution for a 10-7 total dilution 1:10 dilution for a 10-8 total dilution 2. Estimate the concentration of cells in each dilution. Before you can spread a sample on the plate, you need to decide which dilutions are most likely to have a reasonable number of cells so that you will have the appropriate number of colonies. The most statistically valid spread plates should have between 25 and 250 colonies per plate (a colony forming unit—cfu). We will assume that each cell will give rise to one colony. Further, we will estimate that a turbid sample (+++) will contain about 8 x 109 cells per ml, a (++) 8 x 108 cells per ml, and a (+) 8 x 107 cells per ml. Some simple calculations are in order. We will use a turbid sample of (+++) as an example. • If you remove 1 ml of the sample and dilute it in 9 ml diluent (see tube #1), you now have 8 x 109 cells put into a total volume of 10 ml or 8 x 108 cells/ml. • If you remove 0.1 ml from this tube and spread it onto the agar surface, there will be 8 x 107 cells on the plate and each can give rise to one colony. Eighty million colonies on one plate would produce a ‘lawn’ of growth, so this is not a good dilution tube to use. Now do the same type of calculations for the remaining dilutions in the table below and choose three dilutions that you think will contain a reasonable number of cells in a 0.1 and 1 ml volume. Our mixture of three organisms is a (+++) turbidity. Tube # 1 2 3 4 5 6 7 8 Concentration of the inoculum (the starting culture) 8 x 109 8 x 108 Cells/ml after dilution Cells in 0.1 ml 8 x 108 8 x 107 Based on our original estimate of cells per ml, which three tubes are most likely to yield 25-250 colonies on the spread plates (you want to pick one above and below your ideal tube)? Tubes:_____________________ The reason it is important to sample from three different dilutions is because our calculations are based on the fact that the starting culture contains about 8 x 109 cell per ml. By sampling several dilutions, we should obtain at least one plate with a reasonable number of colonies in case our original estimate of cells in our culture is incorrect. 3. Spread the cells on the agar. When all your dilutions are done, you are ready to spread. Aseptically remove 1.0 ml (the table above is using 0.1 ml) from one of the dilution tubes that your calculations show should give a reasonable number of colonies and dispense the liquid onto the surface of one of your TY agar plates made previously and stored in your cabinet. Make sure your lab partner gets a chance to do this spread plate technique. Dispose the pipette tips correctly. a. Take your metal rod that looks like a metal triangle, immerse the triangle end into the ethanol. Place the ethanol coated end into the flame of your Bunsen burner in order to ignite the ethanol (this is fun). Let the ethanol burn off away from the flame (in order to prevent it from getting too hot). Be careful not to let the ethanol run down the handle of the metal triangle or you might burn your fingers. MMBB255 Week 5 Background 5 b. After the ethanol is burned off, allow the rod to cool briefly, place the sterile metal triangle end on the dilution sample you placed on the plate, and spread the liquid all over the surface of the plate. Rub smoothly and evenly over the surface. Place the contaminated metal triangle back into the ethanol for the next plate and if done spreading, flame the stick one last time to decontaminate it and put the stick back were it came from. c. Repeat steps a and b with the other two plates, using a different dilution tube for each plate. Be sure to label each plate with the volume and dilution number of the inoculum. d. Invert the plate and incubate at 30°C until Thursday. C. Fermentations: Wine making and beer continued. 1. The wine is still in the primary fermentation stage. The beer will be bottled Thursday. 2. Check the trap to make sure the seal is intact and fermentation is occurring. You will need to keep doing this until the fermentation is finished. 3. Once no more bubbles are rising in the flask, we will take the final potential alcohol reading and bottle the wine if worthy. Normally the wine is transferred to a new container for another 2 to 4 weeks to finish fermentation and improve flavor (this leaves behind the 'sludge'). We will skip this due to time constraints. This is called the secondary fermentation. To age the wine, it is transferred to a flavoring container (oak barrels are particularly good) and then bottled. Aging for an additional 2 weeks to several years will add complexity and improve the flavor. Most wines today are clarified and sulfites are added to help preserve the wine. We will simply transfer the primary fermentation, trying not to disturb the bottom sludge, to a sterile/clean bottle. Before bottling, the finished product will be analyzed. D. Fermentations: cheese tasting – Finished. At this stage, the cheese should be firm and ready to eat. Homemade cheese is a good substitute for cream cheese in baking. Bacteria, (primarily Lactococcus lactis, Lactococcus cremoris, Streptococcus thermophilus and Lactobacillus bulgaricus) are involved in the early stages of fermentation for most types of cheeses – unripened soft (cottage, cream and mozzarella), ripened (brie), semisoft (blue/Roquefort), and hard ripened cheeses (cheddar, swiss, and parmesan). Following the initial processing (as you just completed) addition of Penicillium sps, Brevibacterium sps, Lactobacillus sps, and Propionibacterium yields ripened cheeses. Taste your cheese. Enjoy! E. Fermentations: sampling the soda – Finished. The soda should be complete by this time. Because the culture has not been allowed to advance to an anaerobic fermentation stage (we stopped it by putting it into the 4°C), no alcohol is produced. Be careful not to disturb the yeast in the sediment when pouring the soda. Although ingestion of the yeast will not harm you, it will spoil the sweet taste of the soda pop. 1. Microscopic examination of the soda pop. Remove a small amount of the sediment and prepare a wet mount. Record your observations at 1000X. wet mount MMBB255 Week 5 Background 6 F. Unknown #2: Isolation & biochemical characterization – Work individually. 50 pts. Due date:_________________ Name: ___________________ Section: __________________ Unknown Number: ________ 1. You will be given a mixture of two organisms in broth (everyone has one red and one non-red one). Record your unknown number above. 2. Streak for isolation. Mix the broth before you streak and label your plate with your name, section, and unknown number. Invert your plate in a bucket that will be incubated at 30°C. After 24-48 hrs, we will grade your technique. Your grade will be based on good isolation of the two different colony types without contamination and good technique. The more isolated colonies you get and use of the whole plate will yield more points. Your streaking grade (15 pts.): _____________________ 3. Identify the two colony morphologies and prepare a Gram stain from each type. Remember to put in all the prerequisite observations for both colony and cellular observations (at 1000 X) to get all points. (18 pts.) Colony type #1 Colony type #2 (Red) Colony characteristics (size, color, texture, smell, etc.) Gram reaction Cell Observations 4. Get a pure culture of your unknown. Use an isolated colony from your streak plate to obtain a pure culture of the non-pigmented (not red) unknown on a second plate. Incubate at 30°C and check your growth after 24-30 hrs. 5. Biochemical tests. Use your pure culture of your unknown to perform the following tests during the biochemical experiment (Week 6). Record your results below. (14 pts.) Sugar fermentation Glucose positive or negative Sucrose Lactose Enzyme production Catalase Oxidase Urease Motility (motile or non-motile) positive or negative 6. Based on the results above and the organisms we have seen so far this semester, I suspect this organism is (write and spell it correctly): (3 pts.) MMBB255 Week 5 Background 7 Thursday’s Procedures A. Fermentations: bottling of the beer. 1. You will analyze the results of the primary fermentation for it’s potential alcohol (using a hydrometer), pH (using pH meter), and any contamination (doing a Gram stain). pH: __________ %Potential Alcohol: ________ Final %Potential Alcohol: ___________ Any contamination (Gram Stain)? 2. It is time to bottle the beer when bubbling ceases. Transfer the primary fermentation to a clean, sterile carboy with a valve. This is what you will analyze in step 1. Add about 3/4 cup (or 70-80 g) of corn sugar dissolved in a small amount of sterile water. This will be the final bit of substrate for the yeast that are still alive and present in the mixture. This final addition of sugar is just enough to generate new growth so that more CO2 will be produced in the bottles - providing the carbonation that is associated with beer. 3. Mix. Using the valve on the carboy transfer the mixture (this is called racking) into sterile bottles to within about 1 inch from the top. 4. Cap the bottles and leave at room temperature for about 1 week. Transfer into refrigeration temperatures for at least a day before testing the final product. B. Technique: the spread plate method for isolation and quantification - continued. Your spread plate should give you two types of information – 1) information about the different types of microbes in your culture and 2) an estimate of the total number of viable cells in the original culture. 1. Identification of colony types. You should see the same three different colony morphologies that you obtained from your practice streak plate that we examined on Tuesday. Compare the two methods. Which method gives the best results? 2. Quantification. Count the number of colonies on the plates (colony forming units or cfu) that fall within 25-250 CFU. Of that total include numbers for each organism (i.e. each colony type). Record this in the table below and then calculate the concentration of viable cells of each organism that were in the original sample by extrapolation. Some terms: a dilution is 1x10-4 or 1:10000; a dilution factor is the inverse of the dilution (i.e. 1/dilution) so 1x104 and 10000. The table below is in the form of this equation: cells of sample/ml = (cfu) x (1/dilution of sample)÷(volume plated in ml) (A hypothetical example is given first). Dilution tube used for spread plate Actual # of colonies x dilution factor ÷volume plated Estimate of cells in the original sample #5 256 1x105 0.1 ml 2.56 x 108 C. Unknown continued. 1. Make sure you do a pure culture of your unknown (see the unknown instructions in Tuesday’s procedures). If you do not have your non-red organism isolated then you must talk with your TA. The Gram stains can be done when you have time as long as you refrigerate your plates. MMBB255 Week 5 Background D. Comparing growth of a bacterium on different substrates – 4 Teams. You will monitor growth of E. coli in two different media and at two different temperatures. You will form 4 teams. During the lab period each team will take 3 time points. At each time point you will take turns within your team (so everyone gets a chance to do all the techniques) to: 1. measure the Abs (OD) at 600 nm with a spectrophotometer. 2. make serial dilutions and pour plates for viable cell counts. The media (medium is singular, media is the plural form) was inoculated in the morning (before lab) with an overnight culture of Escherichia coli by your lab instructor. Please record the zero time. We will be monitoring throughout the day so be careful not to contaminate your medium; use aseptic techniques. •Team A will monitor growth in Luria-Bertani (LB) broth + glucose at room temp (≈22°C). •Team B will monitor growth in Luria-Bertani (LB) broth + glucose at 37°C. •Team C will monitor growth in Luria-Bertani (LB) broth + lactose at room temp. •Team D will monitor growth in Luria-Bertani (LB) broth + lactose at 37°C. Do the following three steps every 30 min (i.e., for the 9:30 lab, you will take a sample at 10:00, 10:30 and 11:00). Each team will take their measurements at each time point in the following order: 1. Take the flask out of the shaker and aseptically remove 5 ml of culture into a sterile test tube and then put the flask back onto the shaker ASAP. 2. One person will aseptically remove 1 ml from the 5 ml sample and make the first serial dilution and pass the remaining 4 ml to their partner who will follow the instruction in step 3. Follow the instructions a – e below to finish the dilutions and plate counts. a. Aseptically remove 1 ml from your 5 ml sample tube and transfer it to a dilution bottle containing 99 ml of sterile diluent. Give the remaining 4 ml to your partner so that s/he can take a spectrophotometer reading. Mix by shaking the bottle in an arc with a paper towel covering the lid (the standard is 25 arcs). This is dilution #1, 1:100 (1/100) or 10-2 total dilution. b. Aseptically transfer 1 ml from dilution #1 to another dilution bottle containing 99 ml of sterile diluent. Mix by shaking the bottle in an arc with a paper towel covering the lid (the standard is 25 arcs). This is dilution #2, 1:100 (1/100) or 10-4 total dilution. c. Aseptically transfer 1 ml from dilution #2 to another dilution bottle containing 99 ml of sterile diluent. Mix by shaking the bottle in an arc with a paper towel covering the lid (the standard is 25 arcs). This is dilution #3, 1:100 (1/100) or 10-6 total dilution. d. Aseptically transfer 1 ml from dilution #3 to a test tube containing 9 ml sterile diluent. Mix with the vortexers or by hand. This is dilution #4, 1:10 (1/10) or 10-7 total dilution. e. Pour plating your cells from dilutions. Before plating your dilution, label the bottom of the plates with the dilution (i.e. 10-7), time point, group, and section. Do the following: • Aseptically place 1 ml from dilution #4 in the center of a sterile petri dish. • Aseptically place 1 ml from dilution #3 in the center of a sterile petri dish. • Aseptically place 1 ml from dilution #2 in the center of a sterile petri dish. Remove a flask of tryptic soy agar (TSA) tempered in the 55°C waterbath. Aseptically pour the TSA to just cover the bottom of one plate and mix the agar with the 1 ml of diluted cells by moving the petri dish gently over the bench forming an figure 8 pattern. Repeat with the other two plates. Allow the plates to solidify on your bench. When the plates have solidified invert and incubate at 37°C for 24 hrs. 3. To measure Absorbance you need to transfer your sample into a spectrophotometer (spec) tube; an optically clear tube that fits in the sample holder – part B in the picture on the next page. Use the same machine for all the time points. The spec tubes we are using need a minimum of 3.5 ml of sample; luckily we have 4 ml left of sample. You will transfer the entire leftover sample into the spec tube; there is no need to use aseptic technique here, just pour the contents in. You will measure the absorbance at 600 nanometers (nm) and record your readings in the table on the next page and in the computer. Follow the following procedures to take your measurements: 8 MMBB255 Week 5 Background 9 Important Notes: 1. Zeroing a machine means to make it read 0. Since a spectrophotometer reads light (detects light) that means you need a 0 %Transmittance (T) or infinity Absorbance (A). Your light path must be blocked for this to occur which means there must be no tubes in the machine. This will correct for any machine error (i.e. leaking light). 2. Blanking a machine means to obtain 100% of the signal (light in our case) which is 100 %T or 0 A. This always should be done with everything that is in the sample except for the thing you are trying to measure (cells in this week’s experiment thus your blank will be LB broth). This will correct for any incident absorbance in the sample that we do not want to measure. 3. These machines have a linear response, i.e. the readings will be directly proportional to amount of analyte (cells), between 0.1 and 1.0 A. If you go over you should dilute your sample (usually a 1:10 will work fine). Procedures for using the Spectronic 20D+ spectrophotometer: a. Turn the machine on with knob A. Let it warm up for 15 min. b. Set the desired wavelength (λ) to desired wavelength using knob C. c. Select the proper filter by flipping the E switch to be in correct wavelength range. d. Zero (see the notes above) the machine by turning knob A until the %T is 0. The reading will flash 1.99 if you try to read absorbance. Use the mode button to see the %T scale for reading. e. Set the mode to absorbance (also called OD for optical density). f. Blank (see the notes above) the machine. Wipe the outside of the blank tube with a Kimwipe and insert it into the sample holder (B). Set the Absorbance to 0 (or 100 %T) using knob D. g. Wipe the outside of the sample tube and insert it into the sample holder (B). Read the absorbance of the sample off the digital readout. h. Record the reading Absorbance readings for Team ________ Time- record real time t= 0 min (we will give you this reading) t=30 min t=60 min t=90 min Absorbance600nm MMBB255 Week 5 Background Study Questions 1. What are the four phases of a growth curve? 2. Can you operate a spectrophotometer? What is the difference between blanking and zeroing the machine? What does this machine detect and which scale is a reading of this detection and which scale is derived from this? 3. Does a spectrophotometer and direct counts measure viable cells? Why? 4. What are some direct cell counting methods? 5. How would you do a viable cell count? What two plating methods could you use to do this? 6. Can you do a dilution problem? Try to answer the dilution problem sets. 7. What is the statistically correct number of colonies for plate counts? 8. Why did we add corn sugar (glucose) to the beer before we bottled it? 9. Can you manually plot growth curve data on a piece of semi-log graph paper and determine the doubling time from the plot? What are the four phases of growth? 10. Define doubling time. 10