Experiment 6 - Reports and Data
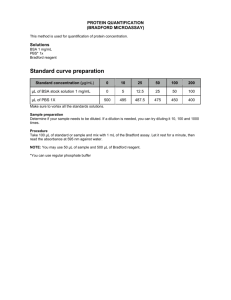
advertisement

MOHAWK COLLEGE OF APPLIED ARTS AND TECHNOLOGY CHEMICAL AND ENVIRONMENTAL TECHNOLOGY DEPARTMENT Lab Report ROOM NO: FE E309 EXPERIMENT NO : 6 TITLE : Total Protein Assay Submitted by : Lyndsay Grover Class : BIOL 10000 lab. Partners : Awatif Hagelamin Instructor : Farag Soliman Date lab performed : March 24th, 2011 Date of submission : March 31st, 2011 FENNELL CAMPUS HAMILTON, ONTARIO 1/6 Purpose: The purpose of this experiment was to use four spectroscopic methods to determine the concentration of a protein solution. The methods used include measurement of the protein’s intrinsic UV absorbance and three methods which generate a protein-dependent colour change: the Lowry assay, the Smith copper/bicinchoninic assay and the Bradford dye assay. This experiment introduce the various assays available to determine protein concentration and also to get us acquainted with the most familiar; the Bradford dye assay. Apparatus: - test tubes - micropipette - cuvettes - Cecil visible (2011) spectrophotometer - reagents: Bradford reagent, BSA stock solution (2mg/L) Safety Guidelines: Reagents may be eye or skin irritant, handle with care. Procedure: 1. Prepare five standard solutions in test tubes containing 0, 250, 500, 1000, and 2000 µg/mL BSA diluted from Protein Standard (BSA) stock solution (2mg/ml) using deionized water. 2. you must set the Cecil visible (2011) spectrophotometer to scan the spectra over a wavelength range from 400 to 700 nm with a speed of 10nm/sec. 3. Use a 4mL plastic cuvette filled with deionized water to blank the spectrophotometer over this wavelength range. Refer to Cecil manual to store baseline reference as preparation for scanning a sample. 4. The prepare five plastic cuvettes containing: 2.0mL Bradford Assay reagent 40µL of protein standard solution from Step 1, to each cuvette starting with the lowest protein concentration and working up, including zero protein. 5. Cover with Parafilm and gently invert several times to mix. 6. Scan the absorbance spectrum of the standards from 400 to 700 nm at 20 nm increments, using an absorbance range of 0 to 2, and a speed of 10nm/sec and note the absorbance at 595 nm. Repeat steps above for remaining protein standards. Print the spectral plots for each solution. 7. Examine the spectra of the standards. If any spectrum has an absorbance at 595 nm greater than 2, dilute the sample by a known amount and repeat the assay. At one wavelength, approximately 575 nm, all of the spectra should have the same absorbance (such an intersection is called an isobestic point and is a defining characteristic of solutions containing the same total concentration of an absorbing species with two possible forms). If any spectrum does not 2/6 intersect the other spectra at or near the isobestic point, it should be adjusted or rejected and repeated. Calculations: 250µL ÷ 1000mL ÷ 2g/mL x 10mL = 1.25mL 500µL ÷ 1000mL ÷ 2g/mL x 10mL = 2.50mL 1000µL ÷ 1000mL ÷ 2g/mL x 10mL = 5.00mL 2000µL ÷ 1000mL ÷ 2g/mL x 10mL = 10.00mL 10mL–1.25mL = 8.75mL 10mL – 2.50mL = 7.5mL 10mL – 5mL = 5mL 10mL – 10mL = 0 Data: Dil. BSA H2O Dilution Abs. 595nm 0 0 10 Dilutions Table 250 500 1.25 2.5 8.75 7.5 1000 5 5 2000 10 0 0 0.43 Absorbance Readings at 595nm 250 500 0.79 0.98 1000 1.39 2000 1.73 3/6 Observations: -BSA clear solution - colour change > BSA with water: 0mL – clear, 1.25mL – clear with light blue tint, 2.5mL – clear light blue, 5mL – clear light blue tint, 10mL – clear blue tint -colour change > dilutions with Bradford: 0mL – gray, 1.25mL – gray with blue tint, 2.5mL – blue with slight gray tint, 5mL – medium blue with very slight gray tint, 10mL – blue - note absorbance at 595nm, scan from 400-700nm at 20nm/sec, absorbance range 0-2 - correct results produce a graph with two humps, the second increasing to the point where it becomes a single hump on the graph Discussion: This experiment is a good introduction for biotechnology students to the process of assays. It gets us familiar with the usage of the Bradford reagent assay which is the most widely used assay due to its efficiency and simplicity. It allows us to observe the patterns that protein concentrations should normally form so that when we perform our own analysis of a singular sample we can use that data to help us understand the concentration of our analyte. This is also a practical experiment for biotechnology students in it familiarizes us with equipment and techniques such as micropipetting and spectrophotometric analysis in order to determine protein concentrations or the concentrations of DNA like in the other experiments we have performed. Sources of Error: Possible Sources of Human Error: Improper micropipette technique. Adding too much or too little of protein standard. Adding too much or too little of Bradford reagent. Insufficient addition of protein samples. Possible Sources of Equipment Error: Spectrophotometer improperly calibrated. Dirty cuvettes. Dirty glassware. Possible Sources of Experiment Error: Improper protein concentration. Improper reagent concentration. Contamination of reagent or protein standard. Post Laboratory Questions: 1. The Bradford reagent assay would be considered non-destructive because it does not require any methods like heating or the addition of an acid in order to break down the compound for a reaction to occur in order for the analysis to take place. 2. This standard plot produced from my data could be used when analyzing samples when trying to find the optimum concentration of an unknown protein to do further analysis and testing on it. Also to predict the rate Bradford reagent will react with their specific protein for analysis based off of the standard data. 4/6 Conclusion: When observing the data and graph and taking into the account the background information provided from the lab I believe we have completed a successful representation of protein standard gradient. The data follows a liner pattern up until the higher concentrations where it begins to curve off as show in the graph. This was discussed in the background information provided in the laboratory manual, describing how the Bradford reagent when acting upon a protein sample will become increasingly non-liner the higher the concentrations. There are no visible signs of errors that seem to be affecting our data as well no data points that would be rejected as they all fit the pattern quite closely. All initial calculations were correct and all data points fall in the absorbance range. I believe this experiment was executed extremely successfully. 5/6 References: Biotechnology, An Introduction S.R. Barnum, 2nd Edition, 2005. Biotechnology 1, Laboratory Manual. Mohawk College, Custom Courseware, 2010. 6/6