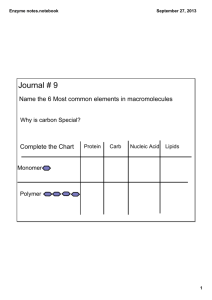
Substrate Channeling
advertisement

METHODS 19, 306 –321 (1999) Article ID meth.1999.0858, available online at http://www.idealibrary.com on Substrate Channeling 1 H. Olin Spivey* ,2 and Judit Ovádi† *Department of Biochemistry and Molecular Biology, 246 NRC, Oklahoma State University, Stillwater, Oklahoma, 74078 –3035; and †Institute of Enzymology, Biological Research Center, Hungarian Academy of Sciences, Budapest, H-1518, P.O.B. 7, Hungary Substrate channeling is the process in which the intermediate produced by one enzyme is transferred to the next enzyme without complete mixing with the bulk phase. This process is equivalent to a microcompartmentation of the intermediate, although classic diffusion occurs simultaneously to varying extents in many of these cases. This microcompartmentation and other factors of channeling provide many potential biological advantages. Extensive examples of channeling can be found in the cited reviews. The choice of methods to detect and characterize substrate channeling depends extensively on the type of enzyme associations involved, the constants of the system, and, to some extent, the mechanism of channeling. Thus it is important to distinguish stable, dynamic, and catalytically induced enzyme associations as well as recognize different mechanisms of substrate channeling. We discuss the principles, experimental details, and limitations and precautions of five rather general methods. These use measurements of transient times, isotope dilution or enhancement, competing reaction effects, enzyme buffering kinetics, and transient-state kinetics. These encompass methods applicable to studies in vitro, in situ, and in vivo. None of these methods is applicable to all systems. They are also susceptible to artifacts without proper attention to precautions. Transient-state kinetic methods clearly excel in elucidating molecular mechanisms of channeling. However, they are often not the best method for initial detection and characterization of the process and they are not applicable to many complex systems. Several other methods that have been successful in indicating substrate channeling are briefly described. © 1999 Academic Press 1 Approved for publication by the Director, Oklahoma Agricultural Experimental Station (OAES). This research was supported in part by NSF Grants MCB-9513613 and BIR-9512912, Oklahoma Center for Advancement of Science and Technology (OCAST) Grant HR98061, and OAES Project OKLO-1393 to H.O.S., and grants from the Hungarian National Science Foundation,OTKA-25291, European Commission Grant INCO-COPERNICUS (ERBIS 15CT960307), MKM-FKFP 158/97, and 1023/97 to J.O. 2 To whom correspondence should be addressed. Fax: (405) 7447799. E-mail: ospivey@Biochem.Okstate.Edu. 306 The term “substrate channeling” designates the coupling of two or more enzymatic reactions in which the common intermediate (I) is transferred from the first enzyme (E 1) to the second (E 2) without escaping into the bulk phase. The simplest of such coupled reactions is shown in Scheme 1. The substrate S is converted to product P via an intermediate I. Substrate channeling has been well documented in vitro, in situ, and in vivo, and with enzymes from prokaryotic and eukaryotic plants and animals [for recent reviews see (1, 2)]. The potential advantages of substrate channeling are more numerous than normally considered. Most of these consequences result from the microcompartmentation of metabolites that is inherent in this mechanism. These potential advantages include: (1) isolating intermediates from competing reactions; (2) circumventing unfavorable equilibria and kinetics imposed by bulkphase metabolite concentrations (3, 4); (3) protecting unstable intermediates (5); (4) conserving the scarce solvation capacity of the cell (6); (5) enhancing catalysis by avoiding unfavorable energetics of desolvating substrates (7); (6) reducing lag transients (times to reach steady-state response to a change in substrate concentration upstream in a coupled reaction path) (8, 9); and (7) providing new means of metabolic regulation by modulation of enzyme associations, e.g., as shown in (10), and increased sensitivities to regulatory signals (11). Thus, studies of substrate channeling are essential for a better understanding of metabolism. Substrate channeling defined operationally as above (intermediate transferred between enzymes without escaping into the bulk phase) can be achieved by several different molecular factors or mechanisms. In fact, many of the examples of substrate channeling probably involve a combination of the basic (elementary) molecular mechanisms, some of which may not be understood or even documented at this time. Therefore, we will avoid a strict definition of mechanisms. However, 1046-2023/99 $30.00 Copyright © 1999 by Academic Press All rights of reproduction in any form reserved. SUBSTRATE CHANNELING the method of detecting and characterizing substrate channeling for an enzyme pair or larger metabolon (12) depends strongly on the nature of the interaction of the enzymes and significantly on the nature by which the intermediate is transferred. Thus, we need to recognize and distinguish among the following processes with which substrate channeling can occur. The best known of these channeling processes transfers covalently attached intermediates from one subunit to another within a stable multisubunit enzyme complex. The oxidative decarboxylation of a-keto acids by pyruvate and a-ketoglutarate dehydrogenase complexes is an example of this general mechanism. We do not discuss methods for characterizing this type of channeling. Channeling of substrates is also now well known for the stable ab complex tryptophan synthase, in which the indole intermediate travels through an internal channel within the enzyme (13). The active sites are 25–30 Å apart, but connected by a molecular tunnel that impedes the escape of indole, which otherwise could escape from the cell since it is an uncharged molecule. On the other hand, just the close proximity of active sites of two enzymes can provide a substantial channeling in addition to the random diffusion path. This is exemplified by enzymes coimmobilized (statically or dynamically) on or in a phase separate from the bulk aqueous phase (14). In principle proximity of active sites might be sufficient for channeling between the sites on soluble multifunctional enzymes, though it is likely that local interactions are also important in retaining at least a portion of the intermediates from escaping into the bulk phase. A specific example of such a process is electrostatic channeling, which uses the favorable electrostatic field between adjacent enzyme sites to constrain a significant fraction of the intermediate within the channeling path. This occurs although the active sites are 40 to 60 Å apart. Brownian dynamic simulations demonstrate that this is much too far apart for channeling by random diffusion mechanisms alone. The correlation of experimental (15) and theoretical (16) studies has established this mechanism especially well. Another process of channeling occurs when the active sites of two enzymes are transiently brought into contact with each other, forming an enclosure that permits direct transfer of the intermediate and sterically prevents its escape into the bulk phase. We believe that NADH channeling between dehydrogenases of opposite chiral specificity for the C-4 hydrogen of NADH is an example of this type (17, 18). The fact that this channeling has never been observed between enzymes of the same chiral specific- SCHEME 1 307 ity forces one to conclude that the NADH does not escape into the bulk phase where its rotation between syn and anti conformations would destroy this chiral specficity in less than a nanosecond. An aspect of great importance in dictating the methods needed for studying substrate channeling is the nature of interaction between the enzyme active sites. Three classes of interactions need to be considered: (1) static (sometimes called stable) complexes, (2) dynamic (sometimes called transient) complexes, and (3) catalytically induced enzyme associations. In contrast to dynamic complexes, the extent of complex formation for static complexes is independent of the enzyme concentrations, at least in the range and conditions of study. Least well known are what we shall call catalytically induced enzyme–macromolecule associations. These are characterized by enzymes (E 2) that exhibit no detectable association with their cognate enzyme (E 1) or protein in the absence of the catalytic reaction. Nevertheless, E 2 associates with E 1 (or E 1– intermediate complex) or a protein as an obligatory part of its catalytic cycle. The case where E 1– intermediate (E 1–I, e.g., E 1–NADH), forms an E 1–I–E 2 complex is equivalent to E 1–I being a substrate of E 2. We know of seven well-documented systems involving either an E 1–I–E 2 or enzyme–protein complex during the catalytic reaction, where, in each case, all attempts to demonstrate an E 1–E 2, an E 1–I–E 2, or a protein–E 2 complex in the absence of the catalytic reaction have failed (5, and references therein, 19). The last two of these seven cases are the acyl-CoA dehydrogenase (a flavoprotein) with the electron tranferring flavoprotein (D. K. Srivastava, personal communication) and several dehydrogenases that channel NADH efficiently, but do not associate at any level of NADH (H. O. Spivey et al., unpublished results). All seven of these systems involve enzymes of very different nature. For many if not all of these seven cases, saturation of the kinetic response falls within the micromolar range. An equilibrium complex with this dissociation constant would be nearly completely associated with 1 mg/ml of enzymes and would be very easy to detect by many physical methods. Such catalytically induced enzyme associations are very similar to binding of the second substrate to an enzyme following a compulsory ordered two-substrate mechanism. That is, no association of the second substrate to the enzyme is detectable in the absence of the reaction. Also the equilibrium dissociation constant has little affect on the K m of the first substrate in a ping–pong mechanism, in sharp contrast to an ordered mechanism (20, p. 143). Knowing these facts should make the catalytically induced enzyme associations easier to accept. We have chosen the word “induced” (associations) rather than “enhanced” to emphasize the fact that enzyme associations in the absence of the catalytic reaction are not experimentally 308 SPIVEY AND OVÁDI detectable. However, the word induced is not meant to preclude small (as yet undetected) extents of association that become greatly enhanced in the catalytic reaction. Thus catalytically induced associations may be considered an extreme example of enzyme associations that are detectable in the absence of the catalytic reaction. This alerts us to the very likely possibility that a catalytic reaction may well alter the extents of the associations between enzymes known to be in dynamic equilibrium, a fact of considerable importance in testing for substrate channeling as we will see. In summary, several different general mechanisms are involved in substrate channeling—first in the manner by which the enzymes interact with each other and second in the mechanisms providing the channeling once the enzymes associate. In addition, the fraction of the reaction flux going by the channeled path is virtually 100% for some of the channeling processes, but significantly less for others. The latter are called “leaky channels.” It is important to know these different processes since the choice of methods for detecting and characterizing substrate channeling depend to a considerable extent on the types of processes involved. The success of a method also depends heavily on the kinetic and equilibrium constants of the enzymes and the conditions of the experiments. Therefore, the best method is very dependent on the system under study; there is no single best method for all systems. Also there are optimal conditions for use of each method, which most often depend on the system constants. We discuss five, more or less general methods in detail: transient-time analysis, isotope dilution or enhancement, competing reaction, enzyme buffering, and, more briefly, transient-state kinetics. A few other methods are mentioned at the end. The relative length of these sections is dictated by how many details were considered important to mention and does not reflect the relative value of a method. under the following conditions or approximations thereto: (1) the reactions are irreversible, (2) the rate of the first reaction is constant over the period recorded, (3) the steady-state concentration of the intermediate [I] ss ! K m2, the K m of the coupling enzyme E 2, and (4) the amount of intermediate bound to the coupling enzyme [E 2–I] is a small fraction of the total intermediate concentration ([I] t 5 [I] 1 [E 2–I]). With these conditions, the rate constant k of the transient phase is the apparent first order rate constant of E 2, i.e., k ; 1/ t 5 k 2 @E2 #/K m2 5 V 2 /K m2, [3] where t is the lag-time for product formation, K m2 and V 2 are the Michaelis constant and limiting velocity of E 2, respectively, and k 2 is the k cat of E 2. The steadystate equations corresponding to Eqs. [1] and [2] are, therefore, @P# ss 5 v 1 ~t 2 t ! and @I# ss 5 v 1 t 5 v 1 K m2/V 2 , @4# which shows that the line representing the steadystate part of the progress curve has intercepts of t and 2[I] ss on the time and [P] axes, respectively (22) (Fig. 1). Equation [3] shows that 1/t is linear with [E 2] for nonassociating enzymes. Physically, the lag time for reactions catalyzed by unassociated enzymes is the time for [I] to increase to 1/e of [I] ss. The rate of the second reaction will increase during the transient state until [I] 5 [I] ss. The first two conditions (irreversible reactions and constant v 1 ) are often well approximated (especially in vitro) by limiting the record to the initial velocity periods of E 1 and E 2, using sufficiently high substrate conditions and using high [E 2] to minimize the reverse reaction of E 1. However, E 1 enzymes with very unfavorable equilibria fail condition 1 and need special treatment (23) as discussed below. Condition 3 METHODS Transient-Time Analysis Principles For the coupled reactions of Scheme 1, where the first reaction proceeds with constant velocity v 1 , the “progress curves,” i.e., time course of formation of P and I, can be represented by (21) P~t! 5 v 1 t 2 @I# 5 v1 ~1 2 e 2kt !, k v1 ~1 2 e 2kt !, k [1] [2] FIG. 1. Transient in coupled enzymatic reactions. Product P versus time showing transient time t 5 10 s and steady-state concentration of I, [I] ss 5 5 mM. SUBSTRATE CHANNELING (“first-order kinetics”) can also frequently be achieved by using sufficiently high [E 2]. Condition 4 (avoiding significant fractions of enzyme-bound I) can often be achieved in vitro, but is frequently not valid in vivo. Also, increasing [E 2] to improve conditions 1 and 3 can create a violation of condition 4 ([I] ' [I] t) as well as raise some practical problems discussed under Other Limitations and Precautions below. Fortunately, even without meeting the above conditions, appropriately modified equations exist for predicting correct transient times. Analytical expressions exist for most of these more complex conditions. However, the most general cases require numerical integration of the model kinetic and conservation equations. Lack of first-order kinetics is easily accommodated if V 2 . v 1 (8, 24), giving t 5 K m2/~V 2 2 v 1 ! [5] @I# ss 5 v 1 K m2/~V 2 2 v 1 !. [6] and Easterby shows that for V 2 /v 1 . 2, the first-order kinetic approximation is adequate, but rapidly loses accuracy at lower ratios (8). By focusing on the intermediate rather than the enzyme, Easterby’s expressions are simpler and valid for more complex conditions (8, 25). This formulation also offers the advantages that integration of rate laws is often not required, and the transient times can be associated with separate physical processes. This approach also provides instructive insights into some general principles. For example, the overall transient time of a sequence of several reactions is the sum of the transient times for each step. These steps include: (a) the transient time to reach the steady state in enzyme forms (free and substrate/product forms of each enzyme), (b) the sum of transient times for each intermediate concentration [I j], and (c) transient time for “feedback” on the pathway (25). The last step allows for variation in the rate of v 1 and includes reversal of the first reaction. The transient times of enzyme forms are typically a few milliseconds or less, whereas transient times of [I j] for most of the applications we discuss in this section are several seconds. Thus for this section, we can ignore the transient times in enzyme forms. The independent extensions of the transition-time analysis of Elcock et al. (16) are referenced here and are discussed further under Experimental Details. Neither Easterby nor Elcock et al. have provided explicit solutions to two additional conditions that are sometimes encountered although Easterby’s general formulations are applicable. The first is the condition where significant fractions of I are bound to E 2 (the enzyme usually in excess over E 1). Several authors have provided solutions to 309 this condition [see references in (23)]. The second condition that is sometimes encountered is one where the first enzyme in the sequence has a very unfavorable equilibrium—the overall reaction proceeds extensively only because of the still more favorable DG of the second enzyme reaction. Yang and Schulz (23) found that their system required this consideration and they provided equations for a two-enzyme sequence to deal with this condition as well as all other conditions 1– 4 enumerated above. Though analytic solutions exist for certain limited conditions, the general solution requires numerical integration. We strongly recommend numerical integration of the model equations whenever there is any doubt about the validity of simpler expressions. An attractive alternative to integrating the quite complex equations of Yang et al. (23, 26) is to use one of the several simulation programs. The easiest of these allows the user to specify the reactions in normal chemical symbols. An editor program translates these reactions into the corresponding differential and algebraic equations and integrates them with user-specified initial conditions (27–30). The Gepasi program offers the advantage that either enzyme kinetic rate laws or rate equations using chemical rate constants can be specified. Enzyme kinetic constants are often known when chemical rate constants are not, and rate equations with enzyme kinetic constants are much more quickly integrated numerically. Equations with enzyme kinetic constants require that steady states of the enzyme forms are established, but these are established within a few milliseconds and are guaranteed to last subsequently throughout the enzyme-catalyzed reactions whenever substrate and intermediate concentrations are much greater than the enzyme concentrations. However, even more general simulation programs can be used. It is easy to learn to write the model equations, which are simply the minimum number of differential equations, each representing the rate law for a reactant plus all the possible conservation of mass equations (this keeps the number of differential equations to a minimum). The set of equations is complete when each variable on the right-hand side of the equations is defined on the left-hand side by another equation. Experimental Details Stable Enzyme Complexes If all the intermediate is transferred directly from E 1 to E 2 in an E 1–I–E 2 complex (transient or stable), the transient time will be zero except for the very fast transient times for enzyme forms (usually less than a few milliseconds). However, within a multienzyme complex some of the intermediate, after dissociating from E 1, may remain in an internal pool that is not mixed with the bulk phase while the remainder of I escapes into the bulk phase (“leaky channeling”). To 310 SPIVEY AND OVÁDI visualize this, one may consider two sequential enzymes that are coimmobilized at high surface density on a membrane. These are often dynamic rather than static complexes in reality, but may be considered stable for the purpose of this illustration. This coimmobilization situation approaches that studied by Goldman and Katchalski (31) and experimentally by Gondo (14, 32). Another example of leaky channeling is electrostatically controlled channeling between associated enzymes (16). With these partial channeling conditions, the experimental transient times will often be less than those of the unassociated enzymes or enzymes without any channeling, but it will not be zero. Ovádi et al. (33) first developed equations for such leaky channeling using the approximation of first-order kinetic response of E 2. Easterby (34) developed equations without this assumption. His analyses demonstrate that the transient time will not be reduced by channeling if: (a) the second enzyme obeys the kinetics of a rapid-equilibrium steady-state mechanism or (b) the coupling enzyme concentration is comparable to its K m or higher. In other cases, a reduction in transient times is predicted. Elcock et al. (16) also developed equations for transient times beyond the “first-order kinetic” condition. Their treatment is based on the probability p c that the intermediate, after release from E 1, is channeled without mixing with the bulk phase, and related probabilities for kinetic events. Their analyses also consider the effects of competitive inhibitors of E 2 and competing reactions (e.g., by E 3) on the transient times. The resulting equations were used to analyze the experimental data from two independent coupled enzyme systems, a natural bifunctional enzyme (35) and a genetically fused enzyme pair (15). Unassociated enzyme pairs were used for reference data sets for both (unassociated enzyme forms for the first study were from a different microbial system in which these enzymes exist as individual monofunctional forms). To use transient times to conclude that substrate channeling is occurring, one must know the kinetic parameters of the associated enzymes, since complex formation can alter these constants from those of the unassociated enzymes. In static complexes where the extent of complex formation is independent of enzyme concentrations, the kinetic parameters of each enzyme can often be determined by addition of only the cosubstrate for the enzyme being characterized. An experimental transient time smaller than predicted by the kinetic parameters of the individual enzymes in the complex is then good evidence of channeling. In addition the extent of channeling a (“channeling efficiency”) can be calculated from these data from all three formulations (16, 33, 34). With an independent method of determining a, e.g., by isotope dilution (see below), the “channeling advantage” b (ratio of internal to external [I]) may also be calculated (34). Analysis is even more definitive if the concentration of the intermediate as well as the product can be measured. These methods have been applied with success to several bifunctional or fusion enzymes (15, 35–37). Dynamic Enzyme Complexes Dynamic enzyme complexes present a more difficult problem for prediction of transient times if the changes in enzyme association alter the kinetic constants. Equations and several different approaches have been developed for this case assuming first-order kinetic behavior of the enzymes and assuming that the catalytic reaction does not alter the extent of enzyme associations (33, 38 – 40). In these cases, the apparent firstorder rate constants, V 2 /K m2, were determined for E 2 in the presence of E 1 at the same concentration as used for measurements of t. This was expected to prevent changes in enzyme associations between these two measurements and any resulting changes in kinetic constants. In addition, Salerno et al. (39) used the dependence of t on [E 2] to include the enzyme associations in their model, and the strategy of increasing [E 2] and [E 1] in constant ratio to maintain [I] ss constant. However, the significant number of catalytically enhanced (virtually induced) enzyme associations now known (see introduction) raises questions about the assumption that use of the same enzyme concentrations during measurement of the apparent first-order rate constant and the t of the coupled reactions guarantees the same extent of enzyme association. Although the E 2 reaction is present in both measurements, the E 1 reaction is present only during measurement of t. This E 1 reaction might alter the extent of enzyme association from that in its absence. This concern can be dismissed if different extents of enzyme association cause smaller changes in the kinetic constants than required to explain the differences in t from that of the nonchanneling reference case. The maximum effects on kinetic constants can be established by saturating E 1 with E 2 (the enzyme normally in molar excess). Also it is conceivable that physical data, e.g., light scattering, could be obtained to test the assumption of changes in enzyme association during the E 2 and E 1 reactions relative to associations with the E 2 reaction alone. Catalytically Induced Enzyme Associations If anomalous (nonclassic) transient times are obtained for catalytically induced enzyme associations, this may be due to substrate channeling, but there are two alternative mechanisms that need to be excluded before this conclusion is rigorous. In one of these, the “association model,” it is proposed that the catalytically induced association changes the enzyme kinetic constants sufficiently to give the greatly shortened t. However, the magnitude of the decreased t and the enzyme SUBSTRATE CHANNELING concentration dependence makes this alternative implausible in some cases. For example, Rudolph and Stubbe (5) found that changing the ratio of E 1 and E 2 gave results that required different kinetic constants for these different progress curves; i.e., the “association model” alone doesn’t fit the data. They also found that it would take a nearly 100-fold lower K m of E 2 in the E 1–E 2 complex relative to the free E 2 to explain the experimental t by a classic diffusion coupling mechanism. Such large changes in the enzyme kinetic constants on complex formation are not likely. Although binding of the protein a-lactalbumin to galactosyl transferase reduces the K m for glucose 700-fold (19), this is a case of the protein complex generating an enzymatic activity that is scarcely present at all in the absence of the protein. It seems unlikely, however that two enzymes that are separately efficient (typically low K m values) would associate to give still 100-fold lower K m values. Also in the case of NADH channeling, increasing free E 1 at constant E 1–NADH actually inhibits the E 2 reaction (17, 41), which is opposite the hypothesis that an E 1–E 2 complex is more efficient (lower K m). The second mechanism for producing anomalous kinetics of coupled reactions that we need to exclude before concluding that substrate channeling exists is what we call the “reactive intermediate” model. This requires no E 1–E 2 associations. It proposes that the intermediate released from E 1 exists initially in a pure highly reactive isomeric form of I for a short lifetime (a few microseconds), which is, however, sufficiently long for this reactive form to diffuse from E 1 to E 2 and be converted to product. This “reactive” form of I has a much higher k cat and/or lower K m than the predominant equilibrium form of I that exists in exogenous samples of I. Isomer equilibrium is established long before I is used from exogenous sources. Such a model was proposed for the apparent anomalous kinetics (low t) of the coupled aspartate aminotransferase (E 1) and malate dehydrogenase (E 2) reactions (42). No E 1–E 2 association could be found between these enzymes at equilibrium (absence of the catalytic reactions). Since catalytically induced enzyme associations were not considered then, the authors proposed this reactive intermediate model as the only other reasonable mechanism for the apparent anomalous t value. Reinvestigation of this system failed to find the experimental anomalies (43), but this is a second potential nonchanneling mechanism that has to be considered and excluded before channeling can be accepted. Fortunately, additional data can be persuasive evidence against this reactive intermediate model. For example, this model still predicts the same type of v-versus-[I] curve as with exogenous intermediate I (usually a Michaelis–Menten curve) although with different K m and V. In the case of putative NADH channeling, such a model does not fit 311 the experimental velocity versus [E 1–NADH] data. Independent of this fact, the K m required by the reactive intermediate model (about 100-fold lower than for free NADH) seems unrealistic for a substrate that already has a low K m relative to other good substrates. In summary, transient-time analysis is a quite rigorous test of substrate channeling for stable enzyme complexes. However, dynamic and catalytically induced enzyme association systems present difficulties that make transient-time analysis less rigorous. Nevertheless, even for these systems, transient-time data can in some cases provide very persuasive indication of channeling when supported by additional data and complementary methods. Other Limitations and Precautions In addition to the limitations discussed above, some systems will have kinetic constants and system constraints that make it impractical to use transient time analysis. This was the case for our study of oxaloacetate channeling in polyethylene glycol-induced solidstate complexes of malate dehydrogenase and citrate synthase (44). Also the time resolution of measurements is limited by the signal amplitude resolution (sensitivity) of the detector (43). For example, despite the 2-ms time resolution of the stopped-flow instrument, in one study the time resolution for the reaction was only 50 ms. This is the result of the fact that very little product is formed during the early part of a transient. Thus longer times are required to reach detectable levels of product formation for longer transient times. Increasing [E 1] improves the time resolution, but raises v 1 and [I] ss of Eq. [4]. If [I] ss approaches K m2, the first-order approximation is lost, but more seriously, eventually v 1 will exceed V 2 whereupon no steady state in I will be achieved. Under first-order rate conditions, the increase in [I] ss can be avoided by increasing E 1 and E 2 in constant ratio. However, increasing [E 2] decreases the experimental t, requiring higher time resolution than before to record it. Higher resolution is provided by the increased [E 1], but by no more than enough to cancel the effect of increased [E 2]. Thus, the inadequate time resolution originally encountered remains. Another precaution to be aware of is that graphical measurements of transient times will almost invariably underestimate the true transient time since v 1 is only asymptotically approached by monitoring P. This error increases greatly as the transient times increase relative to the recorded times. Since the recorded time needs to be limited to prevent product inhibition, etc., this can cause serious errors for the longer transient times. Thus to use progress curves to obtain transient times t, it is far better to obtain v 1 as a best-fit parameter from nonlinear regression analysis than to visually assume its value. The equation to use is either Eq. [1] or its more complex forms depending on the condi- 312 SPIVEY AND OVÁDI tions of the experiments. Nonlinear least-squares analyses can even be done with the numerically integrated equations for the more complex conditions [e.g., (28, 30)]. Isotope Dilution or Enrichment Principles The isotope dilution/enrichment method is the most traditional way to test for substrate channeling. In a sense, this method is a direct test for channeling since the lack of dilution by the bulk phase of the endogenously generated intermediate I is the operational definition of channeling. Scheme 2 illustrates isotope dilution where for simplicity we consider irreversible reactions. An isotopically labeled precursor S* in a sequential path of reactions is added (usually concomitantly) with a large concentration of unlabeled intermediate I that is called the “exogenously” added I. After an appropriate reaction time, one quenches the reactions and isolates a product P. This may be the immediate product of I or one of the products several reaction steps downstream. We will assume nonbranched reactions downstream for simplicity although branched paths can also be dealt with in principle. If the catalytic reactions producing and acting on I are coupled only by classic diffusion of I through the bulk phase between these two enzymes, the labeled (“endogenously produced”) I* will be diluted by the pool of unlabeled I at concentration [I], and the specific radioactivity of P will be reduced relative to that of S*. As illustrated below, the extent of dilution in this case of classically coupled reactions is predictable as a minimum extent if [I*] SS is measured or is reliably predicted. Significantly less dilution of the specific radioactivity than the minimum dilution via the classic diffusion path is evidence of channeling, i.e., is evidence for the absence of equilibration of endogenously generated I* with exogenously added I. In perfect channeling the ratio of specific radioactivities of P* to S* is 1, but it is less than 1 for leaky channeling (channeling efficiencies a less than 1). The same type of information is obtained if the initial substrate is unlabeled and the intermediate is labeled. In this case isotope incorporation would be expected by any nonchanneled path. Experimental Methods The most widely used and generally applicable approach is equally rigorous with all three types of enzyme associations since it does not need to know the SCHEME 2 kinetic parameters of the enzymes involved. That is not to say that the isotope method will work well with all systems. Limitations are discussed below. The procedure used by Lyle et al. (37) to evaluate channeling in the bifunctional enzyme ATP sulfurylase and adenosine 5’-phosphosulfate kinase illustrates the approach. The production of P* is measured during a fixed period for a range of initial concentrations [I 0] of I. From this curve, the [I 0] that reduces the relative specific radioactivity of P*/S* to 50% of that of S* is obtained. In their system, this concentration of exogenous I was 60-fold higher than the endogenously generated [I*] ss. However, the channeling efficiency would be still higher since during the transient phase of [I*], its concentration would be lower than that at its steady state. Therefore, they repeated the experiments, but withheld the exogenous I for a fixed time interval. Enormously higher concentrations of [I 0] were required to give significantly reduced specific radioactivities in P*. The first of these experiments gives a lower limit to the channeling efficiency, which in this case is very high. Specifically for this example, it allows one to say that less than one in 60 molecules escapes the channeling path. If the enzyme kinetic constants and dissociation constant for E 2–I were known, a more quantitative value could be obtained by numerically integrating the model equations using one of the simulation programs described in the section on transient time analysis. These equations would allow for: (1) [I] to be in excess of first-order concentrations; (2) a significant fraction of [I*] binding to E 2; and (3) significant reversal of the E 1 and E 2 reactions. However, this more demanding analysis would probably be justified only when the channeling efficiency was so low as to be in question. Orosz and Ovádi (40) devised a quantitative approach to isotope dilution valid when [I] 1 [I*] are low relative to K m2 (the K m of E 2) and the reactions are irreversible. For the classic, nonchanneling system the specific radioactivity of the product would then be SRAp : @P*# @P*# 1 @P# 5 v 1 t 2 ~v 1 /k! ~1 2 e 2kt ! , v 1 t 2 ~v 1 /k! ~1 2 e 2kt ! 1 @I0#~1 2 e 2kt ! [7] where [I 0] is the initial concentration of the exogenously added I, k 5 V 2 /K m2 and v 2 5 k([I] 1 [I*]). Specific radioactivities significantly larger than predicted by this equation would be evidence of channeling of I between the enzymes. The ratio of specific radioactivities of product to substrate, r 5 SRA P/SRA S, would be 1 for perfect channeling and the lower value predicted by Eq. [7] for nonchanneling. However, the first-order kinetic requirement greatly limits the max- SUBSTRATE CHANNELING imum [I] that can be used to assess channeling, which in turn limits the dilution possible and makes the changes in r small between the channeled and nonchanneled systems. Isotope enrichment experiments might provide larger changes in r. Another problem with this method, unlike the method of Lyle et al. (37), is that it requires knowledge of the kinetic constants K m2 and V 2 . While this is not a problem for static enzyme complexes, it is a problem for dynamic associations and is most likely impractical for catalytically induced enzyme associations for reasons discussed under the transient-time analysis. Studies in Situ In situ systems are especially valuable since they retain much of the complexities of the cell including membranes, cytoskeletal components, and locally high enzyme concentrations. Dilute cell suspensions provide sufficiently slow kinetics to allow valuable experimental procedures despite the high physiological enzyme concentrations. Isotope dilution and enrichment tests are especially good for tests of channeling in situ since they do not require knowledge of the kinetic properties of the enzymes involved. Few other channeling tests are known for these conditions, though some notable exceptions have worked on occasion (45, 46). However, experimental complexities and the precautions required for these radioisotope studies are generally considerably greater than for studies in vitro. In most cases, the cells have to be made permeable to the initial substrate and/or the intermediate. A large number of permeabilizing agents have been used (47–50). Most of these are what we call macropermeabilizing agents, which produce membrane holes larger than the macromolecules within the cell. Surprisingly, loss of enzymes is sufficiently slow in many of these cases to permit the studies desired, which in itself indicates the binding of these enzymes to each other or structural components in the cell. However, a long retention of enzymes is definitely not always found. Fortunately, a small loss of enzymes will not necessarily preclude evidence of channeling, since loss of enzymes would be expected to decrease channeling giving false negative indications. In contrast to macropermeabilizing agents, a-toxin (also known as a-hemolysin), from Staphylococcus aureus, forms a cylindrical heptamer of subunits across the cell plasma membrane with an inner pore 1–2 nm in diameter (51, 52). Molecules smaller than approximately 1000 Da can pass through this pore; however, trypan blue should be excluded (53), contrary to earlier reports. Permeability to trypan blue is thought to result from lytic components contaminating the a-toxin preparation. Commercial preparations of a-toxin are frequently heavily contaminated and have been extremely expensive although this toxin can be easily and safely purified in normal biochemistry laboratories (53). For successful tests of channeling, 313 e.g., by isotope dilution, it is essential that the intermediate have as rapid an access to the same intracellular compartment as the initial substrate. Otherwise, a false-positive indication of channeling can result from lack of the intermediate at the site of metabolism of the substrate. ATP and hexose phosphates were reported to pass freely through a-toxin pores. However, recent studies in progress from several laboratories find that these substrates pass poorly or not at all through these pores. Permeability depends on the cells (54), permeabilizing conditions, and possibly the substrain of S. aureus Woods 46 that is used. These questions on substrate permeability are now under active investigation by a collaboration of laboratories. In addition, the conditions for permeabilization need to be optimized for each cell type for virtually all of the cell permeabilizing methods. Numerous references can be cited for substrate channeling studies with permeabilized cells (55–58). To our knowledge, the most thorough and fullest description of the necessary controls are those of Cohen and coworkers (55) indicating channeling in most of the steps of the urea cycle [see (59) for recent review]. Results of these studies were subsequently fortified with electron microscopic methods demonstrating colocalization of some of the enzymes and their mRNAs (60, 61). Due to variability in cell preparations from experiment to experiment, many of the control experiments must be done with every experiment. Other Limitations and Precautions Numerous practical problems and some limitations need to be understood for successful applications of isotope methods for studies of substrate channeling. The time of quenching of the reaction can be very important, since utilization of exogenous I is proceeding during the reactions. This can be especially important in obtaining definitive values of r when low concentrations of I are used and for studies in situ, where further metabolism of products may complicate the analyses. Isotopically labeled biochemicals are frequently expensive and often not commercially available, requiring one to make them from a labeled precursor. The resulting compounds are not always sufficiently stable for the desired studies. The quenching and subsequent isolation of the product free from isotope contamination have to be guaranteed. This contamination can also come from breakdown of any unstable S* or intermediates—not just the parent compounds. Strong acids or bases are commonly used to quench enzymatic reactions and these promote many decompositions that may contaminate the isolated product. Errors from these sources are especially serious when: (1) the quantity of product is a small fraction of the original substrate (the usual case to maintain a constant velocity) and (2) the quantitative differences in isotope dilutions predicted for different mechanisms 314 SPIVEY AND OVÁDI are small. Thus small decompositions that are not normally thought of can contaminate or alter the radioactivity of the isolated product seriously in these experiments. In some cases, quenching by milder conditions, e.g., with excess EDTA or liquid N 2, can circumvent this problem. CO 2 release from the product is often used to determine specific radioactivities. Despite the long history of such measurements, we have found surprisingly large errors in such determinations due to nonenzymatic decarboxylations and poor retention of CO 2 in the common scintillation cocktails. Recommendations to minimize these are provided by Lehoux et al. (62). Coupled reactions that regenerate the substrate or cofactor, e.g., coupled dehydrogenases or kinases regenerating NAD or ATP, present increased problems since product and substrate are the same and hence cannot be distinguished. In principle, the nonchanneled path will dilute the specific radioactivity of the substrate (product) from its original value, but since the amount of initial substrate is usually in large excess over the regenerated substrate, changes in r will usually be too small for practical use. Isotope enhancement experiments would circumvent this problem to a significant extent, but would still lead to smaller changes in r than in nonregenerating reactions since the unlabeled substrate will dilute the radioactivity of the intermediate that is converted to product/ substrate. This does illustrate the relative advantages of the dilution versus enhancement approaches for certain systems. It is more accurate to compare a small radioactivity against a near-zero background level than to compare radioactivities of similar magnitude. In addition, the labeled intermediate may be more stable or less expensive than the labeled substrate. Isotope dilution experiments have been used to test the “reactive intermediate” model of anomalous kinetics of coupled enzymes. However, the equations proposed to test this model have been disputed (43). It is questionable whether an adequate isotope dilution experiment could be devised to test for this mechanism of coupled reactions. However, independent data can exclude this mechanism or provide supporting evidence for it as discussed under Transient-Time Analysis and under Enzyme Buffering Method sections. Competing Reaction Method Principles and Experimental Details Competing reactions for the intermediate I can provide a very useful indication or test of channeling. This competition can be provided by enzymatic (E 3) or nonenzymatic reactions. One should use a large excess activity of E 3 (or fast nonenzymatic reaction) relative to E 2 to provide a robust test. One assumes or experimentally verifies that E 3 remains in the bulk phase, i.e., it does not bind to the E 1–E 2 complex (but see comments below). The advantages of the test are its experimental simplicity and range of systems to which it is applicable. Like the isotope dilution/enhancement method, it does not require knowledge of any kinetic constants, hence it is applicable in principle to all types of associating systems. It also may be considered a direct test of substrate channeling in the sense that it detects the presence or absence of the intermediate in the bulk phase. We found the competing reaction method especially helpful in demonstrating channeling of oxaloacetate between malate dehydrogenase and citrate synthase in solid-state suspensions of the polyethylene glycol (PEG) induced enzyme complex (44). The experimental difficulties in preparing and maintaining the solidstate enzymes complexes and in mixing such viscous (PEG) solutions make alternative approaches much more difficult if practical at all with this system. This illustrates the dictum that no one method is best for all systems. Subsequently Srere and co-workers have used the competing reaction method with the same E 3 as well as an alternative competing enzyme reaction to test for channeling of oxaloacetate in genetically fused complexes of these enzymes (15, 63). These authors (63) found that increasing [E 3] did not decrease the observed reaction rate (v 2 ) linearly; a decreasing effect at higher [E 3] was found instead. This was initially puzzling, since we had observed essentially complete trapping of oxaloacetate in the solidstate complex of these enzymes (44). However, a simple treatment reveals that the velocity of the competing reaction v 3 } =[E 3] and thus dv 3 /d[E 3 ] } 1/ =[E 3 ], which is qualitatively consistent with the data of Lindbladh et al. The more quantitative model of Elcock et al. (16) also reproduces these data (63). Our ability to virtually abolish the v 2 reaction is most likely due to the nearly 10-fold higher ratio of E 3 activity to that of E 2 in our study (44). With the fused enzyme complex, Lindbladh et al. (63) and Shatalin et al. (15) were also able to apply the transient time method. Competing reactions should decrease transient times. This is most easily understood by realizing that any and all reactions that increase utilization of I will lower [I] ss and hence shorten the time it takes to reach this level via the v 1 reaction. The model equations of Elcock et al. demonstrate this effect more quantitatively (16). It is unclear then why Srere et al. (15, 63) found no changes in the transient times in the presence of the competing reaction. Limitations and Precautions Binding of E 3 to the E 1–E 2 complex or either of its components may decrease the activity of E 3, even eliminating it, but since a large excess of E 3 relative to E 1 and E 2 is used, it is hard to see how this would give a false-positive indication of channeling. Binding of E 3 could either decrease channeling or possibly enhance 315 SUBSTRATE CHANNELING binding of E 1 to E 2, causing a more complex result. Several examples of such cooperative ternary complexes are known, e.g., (64, 65). If this led to increased channeling between E 1 and E 2, this would be an important result though the correct understanding of the observed channeling would require knowing that it was induced by E 3. Appropriate binding data should elucidate this mechanism and use of an alternative competing reaction may be helpful as well. If binding of E 3 to the complex decreased channeling a falsenegative indication could result. In summary, only a possibly false-negative conclusion concerning channeling should result from the competing reaction method if adequate control experiments are made. Reasons for the failure to observe the expected effect of a competing reaction on the transient times (63) remain unclear at this time, but would appear to reside in the experimental method rather than the theory. Enzyme Buffering Method Principles The enzyme buffering method is especially helpful for some systems, but is limited to those where the K d of E 1–I is not more than the K m of E 2, as explained below. The advantages of this method are best exemplified for NADH dehydrogenases where the channeling of NADH is being tested. Therefore, we discuss it in terms of NADH, though it is applicable to some other systems as well. Since the K d of E 1–NAD is usually about 100-fold higher than that of E 1–NADH (66), this method is not practical for testing channeling of NAD in most cases. NADPH channeling has been reported for two different enzyme pairs (67, 68) we know of without a thorough search. NADH channeling presents two special problems. It is an example of an intermediate that is regenerated by E 2 into its original form (NAD). This makes it difficult to apply the methods discussed above. Second, our recent studies to be published elsewhere show that these systems are among the catalytically induced association type; i.e., no association of the two enzymes can be detected in the absence of the catalytic reaction. Thus the test preferred by some of directly measuring the rates of NADH transfer from E 1 to E 2 in the absence of the reaction (69, 70) is not applicable. The enzyme buffering method has been criticized by these authors, but we believe these criticisms are very inaccurate. However, the enzyme buffering method is not a good choice of a channeling test for systems without the special properties of these dehydrogenases (low K d of E 1–I and regeneration of the substrate), and improved methods are needed even for these dehydrogenases for reasons discussed under Further Precautions, Limitations, and Alternative Approaches. The question the enzyme buffering method addresses is equivalent to asking if an E 2 enzyme can use the E 1-bound form (subscript b in text and Scheme 3) of the common intermediate (NADH in our example) in addition to the free form (subscript f). The obvious approach is to replace NADH f with NADH b. This is not entirely possible since these forms are in equilibrium. However, the dissociation constant K d of E 1–NADH is usually about 1 mM, so with excess E 1 it is possible to reduce [NADH] f to a value well below its K m for E 2 (K m2f in Scheme 3). The larger arrow on the left of Scheme 3 indicates that for this binding equilibrium, NADH is normally more than 99% in its E 1-bound form. Thus, the expected (“classic”) velocity, v cl0 5 V 2f/ (1 1 K m2f/[NADH] f), of the E 2 reaction in the presence of high [E 1] can often be reduced to a fraction of the V 2f, the limiting velocity of E 2 for free NADH. Instead the experimental velocity, v exp, is frequently found to be many times v cl0 (3, 17), which is the maximum velocity the reaction could have assuming that E 2 could use only NADH f. The experimental criterion of anomalous kinetics is thus R [ v exp/v cl0. An R value significantly greater than 1 is, therefore, incompatible with the classic mechanism. The most reasonable explanation is that E 2 can use the only other form of NADH that is present in the solution, namely, E 1-bound NADH, which is also the vastly predominant form. The alternative, nonchanneling mechanisms (“enzyme association” and “reactive intermediate” models) discussed under Transient-Time Analysis for such anomalous kinetics do not appear compatible with the data. However, the data do fit the kinetics of a channeling model very well as will be described elsewhere. Thus we consider the R criterion to be a good indication of NADH channeling when R @ 1. NADH channeling has been found only for enzymes of opposite chiral specificity (A or B) for the C-4 hydrogen on the nicotinamide of NADH (18). Thus control experiments using E 1 or E 2 paired with enzymes of the same chiral specificity (3, 17) have demonstrated that the average R is within 11% of 1.00. Thus, the R values of 4 or more found for several enzyme pairs of opposite chirality (3, 17, 41) are 40-fold or more above the reasonable experimental errors. SCHEME 3 316 SPIVEY AND OVÁDI E 1 is not catalyzing any reaction since its cosubstrate is missing. E 1 is simply buffering NADH to a low [NADH] f, hence the name “enzyme buffering test.” In summary, the velocity of E 2 with its cosubstrate is examined in the absence of E 1 and again in the presence of substrate levels of E 1. Data without E 1 give accurate values of K m2f and V 2f and data with varying [E 1] give v exp. v cl0 is calculable for each v exp data point using the K m2f, V 2f, and K d determined in previous experiments. Normal assay concentrations (nanomolar level) of E 2 are used that are typically 10 3- to 10 4-fold more dilute than [E 1]. Thus the dissociation equilibrium of E 1–NADH is not altered by the presence of E 2. Also, the lifetime for equilibration of the E 1–NADH dissociation is a few milliseconds so that this dissociation equilibrium is maintained throughout the steadystate reaction of E 2. Kinetic measurements in the absence of E 1 are almost always well described by the Michaelis–Menten equation, but any other function, e.g., an empirical power series, would serve the purpose equally well. Sigmoidal kinetics, e.g., offer no problem. That is, no kinetic model need be assumed. Experimental Details Typically the kinetic experiments with E 1-buffered NADH are done in two ways: (1) by varying total concentrations of E 1 and NADH, but at constant ratio of [E 1] t/[NADH] t, where subscript t means total, and (2) with constant [NADH] t, but varying [E 1] t (all in excess of [NADH] t). The former provides plots closely approximating the hyperbolic Michaelis–Menten curves representing the E 2 activity with E 1–NADH substrate [see, e.g., (17)]. The latter strategy virtually always gives higher R values in the limit of high [E 1], but the data will not give the usual Michaelis–Menten or double-reciprocal plots. However, combination of both sets of data can be analyzed by nonlinear least-squares methods to obtain all the information contained in the graphical procedures and more. There are primarily only three experimental constants that are needed for this test: K m2f, V 2f, and K d of E 1–NADH. In principle, the molar absorptivity of the E 1-bound rather than free NADH should be used in the calculation. However, the difference between the absorptivities of free and bound NADH are rarely significant relative to other sources of error although we still prefer to measure the absorptivity of E 1-bound NADH. The kinetic constants (K m2f and V 2f), of E 2 for the free NADH are obtained in the usual manner in the absence of E 1. For most dehydrogenases, these constants can be determined with considerable accuracy, e.g., 10% relative standard deviation, since oxidations of NADH are very favorable; i.e., large changes in absorbance occur within the initial velocity portion of the data. The dissociation constant K d can be determined with similar accuracy only with careful attention to correct procedures, which we discuss next. Determination of K d and Related Precautions One of the precautions needed is to ensure that the enzyme is free of bound nucleotides, which can extensively alter enzyme kinetics as well as the NADH binding equilibrium (71). Treatment of the enzyme stock with activated charcoal (Norit SA3 formerly Norit A) has frequently been used to remove bound nucleotides, but Prabhakar et al. (71) claim that their Norit needs further washing to be maximally effective. Ionexchange (72) or dye-affinity chromatography is preferred by some investigators for removing bound nucleotides. These chromatographic methods provide greater recovery of enzymes and may reduce some denaturing effects of charcoal treatment for some enzymes. The absorbance ratio A 280 /A 260 measured before and after treatment is useful to more quickly ascertain the extent of nucleotide binding in future work. Some enzymes have negligible bound nucleotide while others, e.g., mammalian glyceralde-3-phosphate dehydrogenase (GAPD), are normally saturated with bound nucleotide. Apo-GAPD is considerably less stable than the holo-form and hence needs to be used for the K d measurements as soon as possible after preparation. Even then, a fast method of determining K d is often necessary to prevent enzyme denaturation and excessive destruction of NADH through the frequent NADH oxidase activity associated with E 1 preparations. The NADH oxidase problem and ways to reduce it are discussed below. K d has been most frequently determined by fluorescence titrations with E 1 using changes in either NADH or protein fluorescence that accompany the binding of NADH to E 1. This is a fast method permitting titration of a starting enzyme concentration within about 2 min. Stability can also be enhanced by starting titrations with maximum concentrations of NADH and diluting the enzyme and NADH solution successively. Data with several E 1 concentrations are desired. It is best to use nonlinear least-squares fitting to obtain best-fit values of both the binding capacity n (moles of NADH binding sites per mole of protein subunits) as well as K d. Transforming the binding equation to a linear form is not good practice, except for visual inspection, although this practice is entrenched by historical tradition. Transformation to linear form requires a subjective estimate of the signal for enzyme-bound NADH that introduces potentially large systematic errors; it distorts the distribution of random errors, and often mixes the predominant errors into both the dependent and independent variables. Therefore, for the final analysis a nonlinear fit to the normally rectangular hyperbolic binding model is best. The computer program should be capable of giving reliable estimates of the uncertainty in both the n and K d parameters from random errors in the data. Data missing these error estimates are highly questionable. The consequences of SUBSTRATE CHANNELING systematic errors should be considered as well since these can be the predominant source of error and extremely large, e.g., 10 or more times larger than the true K d. This most often happens with data that are confined to near-“stoichiometric” binding, the condition where [E 1] and [NADH] are so high relative to their dissociation constant that almost all of the added limiting component is bound. This is understandable when it is realized that a significant fraction of the limiting component must be free to provide a measure of the strength of binding; otherwise only the maximum capacity (n) of binding is measured. Still another deficiency is that many researchers have calculated K d with the assumption that all the protein binding sites are competent for binding, i.e., binding site concentrations 5 n[E 1], where [E 1] is the subunit concentration. In fact we almost always find the experimental binding capacities to be about 0.6 to 0.7 of the binding site concentrations (calculated from the protein concentrations) for commercial enzymes. The same enzymes purified in our laboratory give n values of 1 within 5% experimental errors, showing that these low binding capacities are not due to our experimental errors. Good channeling can be found with both commercial and purified enzyme preparations, but higher weight concentrations of E 1 giving equivalent NADH binding site concentrations are needed for commercial enzymes to give binding equivalent to that of enzymes with n 5 1. Neglecting this deficiency in binding capacity can give false-positive results. Another potential problem is the presence of cooperative binding of NADH. Surprisingly, the binding isotherms have all been free of this complication in our experience as well as in other published studies, even for enzymes with well-known cooperative binding properties. This is probably because the titration range used covers the binding to predominantly one step in the total ligation range. One needs to be sure that this is the same step that is functioning in the subsequent channeling measurements. Kinetic measurements with a control, NADH acceptor enzyme of the same chiral specificity as E 1 can provide this assurance as described below. The high [E 1] used (10 3–10 5 times higher than normal assay concentrations of enzymes) requires unusually high purity of E 1 with respect to E 2 activity and NADH oxidase activity. Fortunately the presence of these spurious activities are easily detected as described below, if they are considered. Usually there will be NADH oxidation in the presence of E 1, even in the absence of any substrates (“NADH oxidase” activity). Often this can be reduced to sufficiently low levels with purification steps, but not always. In fact this activity is intrinsic to some enzymes, e.g., the lipoamide dehydrogenase component of a-keto-dehydrogenases. This NADH oxidase activity will preclude all but fast methods for K d determinations unless compensated for by 317 NADH regenerating components. Regeneration systems are limited by the normally very unfavorable equilibrium for NAD reduction, but we have found a very good regenerating system that will be described elsewhere. We expect this to greatly improve the accuracy of K d measurements for these systems. Kinetics with E 1 Present To reduce the expense of the high [E 1] used, we use black wall microcuvettes and a solution volume of only 150 ml. Standard spectrophotometers can be adapted easily for these solutions by adding 2-mm-wide horizontal slits to the cell holders. The loss in signal/noise ratios is rarely significant with this reduced light power. E 1 and the components of the reaction mixture are added in sequence to observe changes from each component. E 1 with NADH alone checks for the level of NADH oxidase, and the subsequent addition of the cosubstrate of E 2 checks for the E 2 activity (intrinsic or contaminating) in the E 1 sample. The sum of these represent the background reaction rate. An NADH regenerating system cannot be used for these measurements. Thus, if the background rate is more than 50% of the final rate with E 2 present, it is best to increase the latter rate by using higher [E 2]. This may require using rapid kinetic methods to permit recording the faster rates. However, [E 2] (NADH binding site concentrations) should not be increased to more than 0.1 of [E 1] to prevent distortion of the E 1–NADH equilibrium. We typically use 50 mM [NADH] t and from 200 to 480 mM E 1 binding site concentrations. This assures us that we can get good initial velocities and low [NADH] f. To save on scarce or expensive E 1, it is quite likely that these concentrations can be lowered significantly even with absorption detection and certainly with the higher sensitivity of fluorescence detection. The higher detection sensitivity allows one to stay within the initial velocity range of signal change with the lower NADH concentrations associated with lower [E 1]. However, the essential requirement of this test is to lower the [NADH] f well below K m2f, which requires a high [E 1] t. Many experiments have been done with far too little [E 1] t to achieve this condition, which gives inconclusive results. In practice, it is nearly impossible to obtain a significant test if the K d is larger than the K m2f, even if the predominant flux is through the channeling path, a fact persuasively demonstrated by computer simulations. Yet several authors claim conclusive evidence against channeling with such inadequate experiments (70, 73, 74). Therefore, conditions should be chosen to make K d as low as possible and K m2f as high as possible. Proper choice of pH can make major improvements in meeting this criterion. In principle, addition of competitive inhibitors of E 2 should raise K m2f, although we have not been successful in the few attempts we have made with this approach. 318 SPIVEY AND OVÁDI Data Interpretations The R value is all that is needed to demonstrate the anomalous kinetics of many A–B and B–A dehydrogenase pairs. As described above, the most reasonable interpretation of R significantly greater than 1 is that NADH is directly transferred from one dehydrogenase to another. However, simple kinetic models allow one to calculate the fraction of the reaction flux going through the channel or the classic path (channeling efficiency a as defined above). Data from both strategies (fixed [E 1] t/[NADH] t and fixed [NADH] t) can be combined for nonlinear regression to the model equation. The calculated a values are not sensitive to details of the model. The reason is that the ratio of [NADH] b/[NADH] f is extremely large in well-designed experiments. Also with respect to the ratio of NADH binding sites to NADH t, these experiments are far more physiological than the conventional steady-state enzyme kinetic studies (3, 75). This can also be claimed for many enzyme–substrate systems other than dehydrogenases. Further Precautions, Limitations, and Alternative Approaches During the enzyme buffering measurements, such high [E 1] are used that NADH binding is virtually “stoichiometric.” However, most physical methods for determining K d, including fluorescence, require at least a substantial fraction of the data to be obtained at much lower [E 1] as explained above. This raises the question whether changes in the quarternary structure of E 1 occur between these different concentrations, giving an erroneous K d for the channeling measurements. For example, mammalian glyceraldeyhyde-3phosphate dehydrogenase undergoes concentrationdependent dissociation in this enzyme concentration range. For this and other reasons, we currently prefer to determine K d by pairing the E 1 with an enzyme E 3 of the same chiral specificity. The K m and V of this E 3 are established, which then allows one to calculate [NADH] f from the velocities of the E 3 reaction by rearrangement of the Michaelis–Menten equation. This approach allows one to use precisely the same [E 1] values for determination of [NADH] f as used in the subsequent channeling tests. The resulting data can then be used directly (with a small amount of interpolation of smoothed data if desired) or K d values can be calculated, but it is the calculated [NADH] f that is used in the calculation of R. Commercial A- and B-type enzymes are adequate for these measurements with E 1 in our experience. However, this approach does assume that there is no NADH channeling between enzymes of the same chiral specificity. We have obtained dissociation constants by this method that are quite close to published values when available. However, due to the assumption involved, careful studies should also use independently determined K d values. We also test the effect of the presence of the cosubstrate of E 2 on K d, although we have not found such effects so far. All other components of the buffer that are to be used in the subsequent NADH channeling test are also present. A serious limitation of the enzyme buffering method is that, contrary to early impressions, almost all dehydrogenases have a maximum in the experimental R values. These maxima are dictated by the ratios of enzyme kinetic and equilibrium (K d) constants that are unrelated to the fraction of channeling. Our combined experimental and simulation studies indicate that many A–B dehydrogenases have high NADH channeling even when their R values are close to 1. Thus, the enzyme buffering method provides a robust test when the R criterion is significantly larger than 1, but the test is insensitive in being unable to detect channeling when the kinetic and equilibrium constants are unfavorable. In this regard, it is important to realize that v cl0 is very much higher than the actual velocity going through the free diffusion path if E 1–NADH is a good substrate. This is because the latter substrate is so predominant that little E 2 is left for free NADH to bind to. However, this NADH channeling cannot be presumed to be present, so we have to use the very much higher v cl0 value for the initial test, even if E 1–NADH is a good substrate. Virtues of the enzyme buffering test are its simplicity, speed, presence of good controls for the NADH system, and robustness when R @ 1. Very high R values demonstrating NADH channeling have been found between enzymes from the same or widely different species (3, 17; and our more recent unpublished studies). Nevertheless, cases exist where R values for an enzyme pair are '1 unless these enzymes are from different species. It does not seem reasonable that channeling would exist in the latter case and be absent between the enzymes of the same species. Further support of this view comes from: (1) knowing that R values are constrained for reasons unrelated to channeling ability and (2) computer simulations of model equations and parameters based on real data. Therefore, improved tests of NADH channeling are needed, even if they are more complex. We are exploring such possibilities, focusing initially on fluorescence polarization measurements on E 2 during the catalytic cycle. Transient-State Kinetics of Enzyme Forms (Fast Kinetics) Rapid kinetic measurements with substrate level enzyme concentrations, e.g., 10 mM to several hundred micromolar, can in principle directly detect enzyme forms and their transitions during the reaction mechanism. Thus, fast kinetic measurements are capable of providing more insight into molecular mechanisms than steady-state methods, which can normally detect only transitory enzyme forms capable of addition or SUBSTRATE CHANNELING release of substrates or products. When detectable spectral changes occur during transitions of enzyme forms, continuous monitoring of such transitions in time is possible. Furthermore, time-resolved wavelength spectra are now possible, e.g., 300- to 500-nm spectra every few milliseconds, and provide the most insight in these circumstances. When spectral changes are not observable, rapid quenching of the reactions is often possible, providing fast kinetic data for a wider range of applications. There are practical problems that often limit the number of elementary steps that can be determined, but fast kinetic methods can provide unique insights into enzymatic mechanisms. Recent microcapillary mixing devices made by microfabrication techniques have achieved 110-ms (76) and less than 10-ms (77) mixing times. These provide valuable practical advances extending kinetic studies to faster elementary processes than previously possible and offering large economy of reagents. Good reviews of transient-state methods have been published by Johnson (78, 79), among others. However, we mention below several practical limitations of and precautions with these methods that are not discussed in the cited reviews. Impressive examples of spectral studies of substrate channeling are those of Dunn and co-workers (13). When rapid quench methods are used, it is often assumed that all elementary steps of the reaction sequence are quenched as rapidly as the overall production of product. This is not a safe assumption. For example, it has been shown by spectroscopic monitoring that reactions of important intermediates proceed considerably longer than the quenching of the overall reaction (13). This oversight from rapid quenching data alone caused erroneous conclusions concerning the basic mechanism of the reaction. Without such independent methods to test for these more slowlyquenched steps, it is not clear how such artifacts can be detected with quench flow kinetic methods. It is also necessary to ensure that the quenching conditions do not decompose the reaction intermediates that are being characterized. Another precaution is to realize that most simulated fits are not unique solutions. Therefore, overlapping and independent data are critical in reducing the number of alternative fits. A recent study is illustrative of chemical quench methods (80, 81). Despite the value of transient-state kinetics for providing details of the molecular mechanisms, they are not in general the methods of choice for initial detection and characterization of channeling. Furthermore, their applications appear limited to highly purified systems, which excludes, e.g., studies of channeling in situ. Other Channeling Tests A variety of other methods have been used to indicate substrate channeling. Especially intriguing are those indicating channeling in vivo. In one method, 319 asymmetrically labeled substrate is added, which is transformed through symmetrical intermediates, yet asymmetrically labeled products are found (45). Another method used 31P NMR magnetization transfer measurements to indicate the channeling of ATP between the pyruvate and creatine kinases in free solution, much like the NADH channeling discussed above (82). Unfortunately, the method is not currently practical for isotopes other than 31P due to either lack of sensitivity or insufficient changes in chemical shifts. A rather common indication of channeling is the finding that precursors are used considerably faster or exclusively in preference to intermediates in a path producing the product (57, 83, 84). Another study found that [ 13C]acetoacetyl-CoA was incorporated into hydroxymethylglutaryl-CoA without mixing with the acetate pool (85). A novel approach using inhibitors indicated in vivo channeling of substrates for b-carotene biosynthesis (46). Still another study used the fact that 13Clabeled intermediates did not escape from permeabilized vascular smooth muscle as evidence that channeling occurs in the glycolytic path of these cells (86). The value of computer modeling, especially when correlated with experimental work, has been demonstrated by the elegant studies of Elcock et al. (16). The prevalence and potential significance of substrate channeling should motivate the development of improved methods. ACKNOWLEDGMENTS We thank John S. Easterby, Adrian H. Elcock, Michael F. Dunn, Eric A. Lehoux, and Leonard A. Fahien for their helpful discussion and recommendations on this manuscript. Note added in proof. Further literature search revealed that a heteroprotein complex can be observed between galactosyltransferase and a-lactalbumin in the presence of the first three of the four substrates: Mn 21, UPD-galactose, and a-lactalbumin and the product UDP. [Brew, K., and Powell, J. T. (1976) Fed. Proceedings 35, 1892– 1898), i.e., glucose need not be present. Thus, Ref. (19) in our review is not an example of a heteroprotein complex that is observable only during the catalytic reaction. However, it remains an example of the special conditions and conformation of the proteins required for this complex formation. Table 6 in this reference by Brew and Powell contains a typographical error. The K d’s summarized should be mM rather than mM units as both practical considerations and an original manuscript of one of the authors indicate. REFERENCES 1. Ovádi, J. (1995) Cell Architecture and Metabolic Channeling, Molecular Biology Intelligence Unit, R. G. Landes Co., Austin, TX. 2. Agius, L., and Sherratt, H. S. A. (Eds.) (1997) Channelling in Intermediary Metabolism, Portland Press, London. 3. Ushiroyama, T., Fukushima, T., Styre, J. D., and Spivey, H. O. (1992) in From Metabolite, to Metabolism, to Metabolon (Stadt- 320 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. 22. 23. 24. 25. 26. 27. 28. 29. 30. 31. 32. 33. 34. 35. SPIVEY AND OVÁDI man, E. R., and Chock, P. B., Eds.), Curr. Top. Cell. Regul. Vol. 33, pp. 291–307. Academic Press, New York. Ovádi, J., Huang, Y., and Spivey, H. O. (1994) J. Mol. Recog. 7, 265–272. Rudolph, J., and Stubbe, J. (1995) Biochemistry 34, 2241–2250. Atkinson, D. E. (1969) in Current Topics in Cellular Regulation (Horecker, B. L., and Stadtman, E. R., Eds.) Vol. 1, pp. 29 – 43, Academic Press, New York. Dewar, M. J. S., and Storch, D. M. (1985) Proc. Natl. Acad. Sci. USA 82, 2225–2229. Easterby, J. S. (1981) Biochem. J. 199, 155–161. Westerhoff, H. V., and Welch, G. R. (1992) in From Metabolite, to Metabolism, to Metabolon (Stadtman, E. R., and Chock, P. B., Eds.), Curr. Top. Cell. Regul. Vol. 33, pp. 361–390, Academic Press, New York. Beeckmans, S., Van Driessche, E., and Kanarek, L. (1990) in Structural and Organizational Aspects of Metabolic Regulation (Srere, P. A., Jones, M. E., and Mathews, C. K., Eds.), UCLA Symposia on Molecular and Cellular Biology, New Series, Vol. 133, pp. 245–257, Alan R. Liss, New York. Kholodenko, B. N., Demin, O. V., and Westerhoff, H. V. (1993) FEBS 320, 75–78. Srere, P. A. (1985) Trends Biochem. Sci. 10, 109 –110. Brzovic, P. S., Ngo, K., and Dunn, M. F. (1992) Biochemistry 31, 3831–3839. Siegbahn, N., Mosbach, K., and Welch, G. R. (1985) in Organized Multienzyme Systems (Welch, G. R., Ed.), pp. 271–301, Academic Press, New York. Shatalin, K., Lebreton, S., Rault-Leonardon, M., Velot, C., and Srere, P. A. (1999) Biochemistry 38, 881– 889. Elcock, A. H., Huber, G. A., and McCammon, J. A. (1997) Biochemistry 36, 16049 –16058. Srivastava, D. K., and Bernhard, S. A. (1985) Biochemistry 24, 623– 628. Srivastava, D. K., Bernhard, S. A., Langridge, R., and McClarin, J. A. (1985) Biochemistry 24, 629 – 635. Ebner, K. E., and Magee, S. C. (1975) in Subunit Enzymes: Biochemistry and Functions (Ebner, K. E., Ed.), Vol. 2, pp. 137– 179, Marcel Dekker, New York. Cornish-Bowden. (1995) Fundamentals of Enzyme Kinetics, 2nd ed., Portland Press, London. McClure, W. R. (1969) Biochemistry 8, 2782–2786. Easterby, J. S. (1973) Biochim. Biophys. Acta 293, 552–558. Yang, S.-Y., and Schulz, H. (1987) Biochemistry 26, 5579 –5584. Storer, A. C., and Cornish-Bowden, A. (1974) Biochem. J. 141, 205–209. Easterby, J. S. (1986) Biochem. J. 233, 871– 875. Yang, S. Y., Cuebas, D., and Schulz, H. (1986) J. Biol. Chem. 261, 15390 –15395. Mendes, P. (1997) Trends Biochem. Sci. 22, 361–363. Mendes, P., and Kell, D. B. (1998) Bioinformatics 14, 869 – 883. Kuzmic, P. (1996) Anal. Biochem. 237, 260 –273. Frieden, C. (1993) Trends Biochem. Sci. 18, 58 – 60. Goldman, R., and Katchalski, E. (1971) J. Theor. Biol. 32, 243– 257. Gondo, S. (1977) Chem. Eng. J. 13, 153–163. Ovádi, J., Tompa, P., Vértessy, B., Orosz, F., Keleti, T., and Welch, G. R. (1989) Biochem. J. 257, 187–190. Easterby, J. S. (1989) Biochem. J. 264, 605– 607. Trujillo, M., Donald, R. G. K., Roos, D. S., Greene, P. J., and Santi, D. V. (1996) Biochemistry 35, 6366 – 6374. 36. Penverne, B., Belkaı̈d, M., and Hervé, G. (1994) Arch. Biochem. Biophys. 309, 85–93. 37. Lyle, S., Ozeran, J. D., Stanczak, J., Westley, J., and Schwartz, N. B. (1994) Biochemistry 33, 6822– 6827. 38. Ovádi, J., and Keleti, T. (1978) Eur. J. Biochem. 85, 157–161. 39. Salerno, C., Ovádi, J., Keleti, T., and Fasella, P. (1982) Eur. J. Biochem. 121, 511–517. 40. Orosz, F., and Ovádi, J. (1987) Biochim. Biophys. Acta 915, 53–59. 41. Srivastava, D. K., Smolen, P., Betts, G. F., Fukushima, T., Spivey, H. O., and Bernhard, S. A. (1989) Proc. Natl. Acad. Sci. USA 86, 6464 – 6468. 42. Bryce, C. F. A., Williams, D. C., John, R. A. L., and Fasella, P. (1976) Biochem. J. 153, 571–577. 43. Manley, E. R., Webster, T. A., and Spivey, H. O. (1980) Arch. Biochem. Biophys. 205, 380 –387. 44. Datta, A., Merz, J. M., and Spivey, H. O. (1985) J. Biol. Chem. 260, 15008 –15012. 45. Srere, P. A., Sherry, A. D., Malloy, C. R., and Sumegi, B. (1997) in Channelling in Intermediary Metabolism (Agius, L., and Sherratt, H. S. A., Eds.), pp. 201–217, Portland Press, Miami, FL. 46. Candau, R., Bejarano, E. R., and Cerdá-Olmedo, E. (1991) Proc. Natl. Acad. Sci. USA 88, 4936 – 4940. 47. Felix, H. (1982) Anal. Biochem. 120, 211–234. 48. Ahnert-Hilger, G., G., Bhakdi, S., and Gratzl, M. (1985) J. Biol. Chem. 260, 12730 –12734. 49. Knight, D. E., and Scrutton, M. C. (1986) Biochem. J. 234, 497–506. 50. Hapala, I. (1997) Crit. Rev. Biotech 17, 105–122. 51. Song, L., Hobaugh, M. R., Shustak, C., Cheley, S., Bayley, H., and Gouaux, J. E. (1996) Science 274, 1859 –1866. 52. Ellis, M. J., and Hebert, H. (1997) J. Struct. Biol. 118, 178 – 188. 53. Hohman, R. J. (1988) Proc. Natl. Acad. Sci. USA 85, 1624 – 1628. 54. Bhakdi, S., Weller, U., Walev, I., Martin, E., Jonas, D., and Palmer, M. (1993) Med. Microbiol. Immunol. 182, 167–175. 55. Cheung, C.-W., Cohen, N. S., and Raijman, L. (1989) J. Biol. Chem. 264, 4038 – 4044. 56. Clegg, J. S., and Jackson, S. A. (1990) Arch. Biochem. Biophys. 278, 452– 460. 57. Stapulionis, R., and Deutscher, M. P. (1995) Proc. Natl. Acad. Sci. USA 92, 7158 –7161. 58. Bladergroen, B. A., Geelen, M. J. H., Reddy, A. C. P., Declercq, P. E., and van-Golde, L. M. G. (1998) Biochem. J. 334, 511– 517. 59. Cohen, N. S., Cheung, C.-W., and Raijman, L. (1997) in Channelling in Intermediary Metabolism (Agius, L., and Sherratt, H. S. A., Eds.), pp. 183–199, Portland Press, London. 60. Cohen, N. S., and Kuda, A. (1996) J. Cell. Biochem. 60, 334 – 340. 61. Cohen, N. S. (1996) J. Cell. Biochem. 61, 81–96. 62. Lehoux, E. A., Svedružić, Ž., and Spivey, H. O. (1997) Anal. Biochem. 253, 190 –195. 63. Lindbladh, C., Rault, M., Hagglund, C., Small, W. C., Mosbach, K., Bülow, L., Evans, C., and Srere, P. A. (1994) Biochemistry 33, 11692–11698. 64. Fahien, L. A., and Teller, J. K. (1992) J. Biol. Chem. 267, 10411– 10422. 65. Knull, H. R. (1990) in Structural and Organizational Aspects of Metabolic Regulation (Srere, P. A., Jones, M. E., and Mathews, SUBSTRATE CHANNELING 66. 67. 68. 69. 70. 71. 72. 73. 74. 75. C. K., Eds.), UCLA Symposia on Molecular and Cellular Biology, New Series, Vol. 133, pp. 215–228, Wiley–Liss, New York. Dalziel, K. (1975) in The Enzymes (Boyer, P. D., Ed.), Vol. 11, 3rd ed., pp. 1– 60, Academic Press, New York. Srivastava, D. K., and Bernhard, S. A. (1987) Annu. Rev. Biophys. Biophys. Chem. 16, 175–204. Fahien, L. A., Laboy, J. I., Din, Z. Z., Prabhakar, P., Budker, T., and Chobanian, M. (1999) Arch. Biochem. Biophys., 364, 185–194. Wu, X., Gutfreund, H., Lakatos, S., and Chock, P. B. (1991) Proc. Natl. Acad. Sci. USA 88, 497–501. Arias, W. M., Pettersson, H., and Pettersson, G. (1998) Biochim. Biophys. Acta 1385, 149 –156. Prabhakar, P., Laboy, J. I., Wang, J., Budker, T., Din, Z. Z., Chobanian, M., and Fahien, L. A. (1998) Arch. Biochem. Biophys. 360, 195–205. Bloch, W. A., MacQuarrie, R. A., and Bernhard, S. A. (1971) J. Biol. Chem. 246, 780 –790. Chock, P. B., and Gutfreund, H. (1988) Proc. Natl. Acad. Sci. USA 85, 8870 – 8874. Brooks, S. P. J., and Storey, K. B. (1991) Biochem. J. 278, 875– 881. Fahien, L. A., and Chobanian, M. C. (1997) in Channeling in Intermediary Metabolism (Agius, L., and Sherratt, H. S. A., Eds.), pp. 219 –236, Portland Press, London. 321 76. Bökenkamp, D., Desai, A., Yang, X., Tai, Y.-C., Marzluff, E. M., and Mayo, S. L. (1998) Anal. Chem. 70, 232–236. 77. Knight, J. B., Vishwanath, A., Brody, J. P., and Austin, R. H. (1998) Phys. Rev. Lett. 80, 3863–3866. 78. Johnson, K. A. (1992) in The Enzymes (Sigman, D., Ed.), Vol. 20, 3rd ed., pp. 1– 61, Academic Press, New York. 79. Johnson, K. A. (1995) Methods Enzymol. 249, 38 – 61. 80. Liang, P.-H., and Anderson, K. S. (1998) Biochemistry 37, 12195–12205. 81. Liang, P.-H., and Anderson, K. S. (1998) Biochemistry 37, 12206 –12212. 82. Dillon, P. F., and Clark, J. F. (1990) J. Theor. Biol. 143, 275–284. 83. Prem veer Reddy, G., and Pardee, A. B. (1980) Proc. Natl. Acad. Sci. USA 77, 3312–3316. 84. George, T. P., Morash, S. C., Cook, H. W., Byers, D. M., Palmer, F. B., and Spence, M. W. (1989) Biochim. Biophys. Acta 1004, 283–291. 85. Miziorko, H. M., Laib, F. E., and Behnke, C. E. (1990) J. Biol. Chem. 265, 9606 –9609. 86. Hardin, C. D., and Finder, D. R. (1998) Am. J. Physiol. 274, C88 –C96.