Biology Lab Report - Sites at Penn State
advertisement

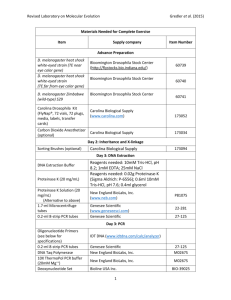
Use of Drosophila Melanogaster as a Model System in the Study of Human SodiumDependent Multivitamin Transporter Michael Brinton BIOL 230W.001 28 October 2013 TA: Sashi Gollapudi Introduction Many human diseases are caused at the genomic level by mutations in DNA coding. These mutations cause defecting proteins to be built, which can have devastating effects downstream. The genomic aspect to human disease can be difficult to study for many reasons, including the lack of family genomic history (due to lifespan) and limited number of progeny on top of the possible danger for the human subjects. Because of this, researchers have developed model systems in order to study human disease. One of these models is the fruit fly, Drosophila melanogaster. Fortunately, proteins tend to be made up of protein domains. These domains are functional groups of amino acids that tend to retain their function regardless of where they appear. They tend to be highly conserved through evolution due to their specialized function in the cell. Thus, they appear in many organisms. In fact 90% of protein domains are replicated in all eukaryotic organisms. This allows for there to be a great amount of homology between certain organisms. Drosophila and Homo sapiens are two of these organisms. The study of drosophila a homolog has been made easier by the production of cDNA libraries. (1) cDNA is copy DNA or complimentary DNA. It is a copy of the organism’s original DNA, made by reverse transcription of mRNA back to DNA. This reaction is catalyzed by reverse transcriptase. cDNA is, essentially, a copy of the organism’s DNA. However, since the cDNA is made from mature mRNA, it lacks the introns that may have been present in the original DNA. Thus, cDNA only contains coding regions of DNA. This makes genes readily identifiable, and can simplify the study of genes. Collections of cDNA can be made and placed into a host cell. This collection is called a cDNA library. Together the host cells containing the cDNA libraries constitute a portion if the original organism’s transcriptome. Creating cDNA libraries in cells like bacteria can make replication and collection of the cDNA libraries for study much easier. In this experiment, cDNA libraries from Drosophila melanogaster will be used within E. coli cells to start. The plasmids containing the cDNA will be isolated and the cDNA will be sequenced and analyzed. This drosophila cDNA will be compared against the human genome to attempt to find a human homolog to the genes of drosophila, thus, showing the importance of drosophila as a model system for human disease. Methods and Materials Taken from: “cDNA Isolation and Analysis: Identifying potential Drosophila melanogaster models of human disease.” Edited by Nelson, K. and Burpee, D. Department of Biology, The Pennsylvania State University, University Park, PA. (2013) Week 1 - Bacteria Culture Selection and Growth: E. coli containing a plasmid with a gene for ampicillin resistance and a Drosophila cDNA sequence were grown on an agar plate. From this, two colonies were selected to continue in the process. Each of these colonies was placed in a glass culture tube containing 2 ml of Luria broth plus ampicillin. These cultures were grown overnight at 37 degrees Celsius. After growing, the cultures were stored at 4 degrees C for six days. Week 2 - DNA Isolation: Plasmid DNA was isolated using the QuickLyse Miniprep Plasmid DNA purification system. Cells were removed from the liquid broth and were resuspended in a lysis solution. Cells were lysed and, using a centrifuge for 1 minute at 13,000 rpm, DNA was captured on the membrane of the spin column. This DNA was then washed in an isopropanol buffer and then eluted in a low-salt buffer. This product would be used in the following steps. Week 2 - PCR Setup: A small portion of each plasmid DNA was set up for a PCR reaction. Each PCR tube was filled with 24 µl of the PCR master mix (containing the buffer, dNTPs, two primers and the Taq polymerase). Added to these tubes was 1 µl of DNA A, DNA B, or sterile water. These tubes were labeled 5A, 5B, and 5N. The contents of these tubes can also be seen in Table 1. Table 1. PCR Tube Contents Tube A Tube B Tube Neg Master Mix 24 µl 24 µl 24 µl Plasmid DNA A 1 µl --- --- Plasmid DNA B --- 1 µl --- Sterile Water --- --- 1 µl These were then submitted to the Teaching Assistant for PCR processing. These samples were then stored in the freezer until week 3. Week 3 - Agarose Gel Electrophoresis: The agarose gel was made by mixing 250 mg of agarose with 25 ml of 1XTAE buffer and microwaving for 40 seconds. 1 µl of ethidium bromide was added to this mixture before casting. Once the gel was cooled and solid, the electrophoresis unit was filled with 1XTAE buffer. Once casted, with 8 wells, the following were prepared. 2 µl of each of the Plasmid DNAs were mixed separately with 8 µl sterile water and 2 µl of 6x loading dye (Bromophenol blue and Xylene cyanol). 4 µl of 6x loading dye was added to each of the PCR tubes from the previous week. The wells of the gel were then loaded as follows. Well 1, 5 µl DNA ladder. Well 2, 12 µl Plasmid A DNA. Well 3, 12 µl Plasmid A PCR. Well 4, 12 µl Plasmid B DNA. Well 5, 12 µl Plasmid B PCR. Well 6, PCR Negative Control. Electrophoresis was run at 107 Volts until the dye had migrated approximately halfway across the gel. The gel was removed from the unit and photographed under UV light. After viewing the photograph and confirming the presence of usable DNA, these sampled were submitted for sequencing at the nucleic acid facility on campus. Sequencing was performed using Sanger’s dideoxy method, and results from sequencing were reported back in ab1 files. Week 4 - Sequence Analysis: The .ab1 file provided by the sequencing facility was imported into MEGA for analysis. A sequence of approximately 450 base pairs was selected starting around base 70 of the provided sequence data. This selection was used to run a nucleotide Blast search. Accession numbers for the drosophila gene and protein and the human homolog gene and protein were retrieved. These were used in cDART (an NCBI resource) to retrieve information about the protein structure and function. A literature search was run using NCBI PubMed to retrieve more information regarding these proteins. Results When bacteria colonies were observed at the beginning of week two, one could see a pellet at the bottom of the tube. The bacteria that had grown overnight had clumped at the bottom of the tube. After isolation of the DNA and further preparation, the samples were ready for PCR. PCR and agarose gel electrophoresis were performed to acquire the gel in Figure 1. Figure 1. Agarose Gel with Plasmid DNA and PCR In Figure 1, the left-most lane (lane 1) contains the DNA ladder. Lanes 2 and 4 contain Plasmid A and B DNA respectively, and lanes 3 and 5 contain Plasmid A and B PCR respectively. Lane 6 contains the PCR negative control. As one can see, both the DNA and the PCR contain DNA of relatively large size. Also, the negative control shows no DNA, which confirms no contamination of the samples. After confirmation of DNA presence, the Plasmid DNA samples were submitted for sequencing as described above. The Blast search revealed that our sequence was drosophila gene NM_137621.2 which codes for drosophila protein NP_523800.1. This is homologous to human gene NM_021095.2 which codes for human protein NP_066918.2. This protein is a sodium-dependent multivitamin transporter. This protein consists of only one domain, SLL5-6-like sbd solute carrier, solute binding domain. This domain is a sodium/glucose transporter. It also functions as a sodium and calcium dependent neurotransmitter transporter. It consists of functional core of 10 transmembrane helices, and functions mainly as a transmembrane transporter. Discussion This experiment ended with the acquisition of information on a drosophila gene and its human homolog. These genes are very similar and code for almost-identical proteins. These proteins contain the same domain. As mentioned above, the gene isolated from the cDNA library was a gene to code a sodium-dependent multivitamin transporter. This protein is responsible for the uptake of certain vitamins into the cell. Many research studies, including those cited below, focus on the function of the sodium dependent multivitamin transporter in the uptake of biotin. Research has shown that this protein may play a part in some diseases. For example a study by Patel showed that there was s higher expression of the sodium-dependent multivitamin transporter (SMVT) in breast cancer (T47D) cells than in normal mammary epithelial (MCF12A) cells. The SMVT is an important carrier-mediated system in the uptake of biotin into the cells. In this study, they found that the cancer cells were able to uptake more vitamins and more biotin because of the higher expression of this gene. This led the researchers to the possibility of using this gene and/or protein in the study of biotin-conjugated anti-cancer drugs and drug delivery systems. (4) Another study focused on the role of the sodium-dependent multivitamin transporter in the uptake of biotin in three different types of cells. In this study, they confirmed the presence of SMVT and its role in the uptake and transport of biotin and the conjugate in MDCK-MDR1 cells. This study led the researchers to use these MDCK-MDR1 cells in the study of the permeability of biotin conjugated prodrugs. The goal is to study the use of drugs such as HIV protease. This study shows again that the SMVT can potentially be used to make drugs targeting specific cell, in this case cells that cause an otherwise difficult to treat disease. (2) A third study also investigated the role of the sodium-dependent multivitamin transporter to enhance the permeability of some drugs and the targeting of specific cells. In this case, they used human-derived prostate cancer (PC-3) cells. This study also focused on the uptake of biotin in these cells. The study also investigated the regulatory pathways for the uptake of biotin. In the end, the study confirmed the expression of the sodium-dependent multivitamin transporter and its functionality within the PC-3 cells. This again led the researchers to the possibility of using the SMVT as an agent in the treatment of prostate cancer. (3) As one can see, the sodium-dependent multivitamin transporter has the potential to be targeted as the transporter for otherwise poorly-permeable drugs. All of the studies above use some sort of model system. In this experiment, we studied drosophila melanogaster as a potential model system for human disease. The fact that we were able to isolate and sequence drosophila DNA from the plasmids in E. coli shows that drosophila can be potentially used as a model system. Finding a human homologue for the isolated gene that was almost identical to the Drosophila gene confirms that Drosophila could be used as a model system. The studies mentioned above show the importance of using model systems. In none of these studied did they use a human test subject. Instead, they scaled down their experiments by using a model. None of these studies happened to use drosophila as the model; however, this experiment shown the possibility of doing so. The drosophila model could have significance in molecular research. Just as the drosophila SMVT gene was very similar to a human gene, so are many of its other genes. The drosophila model is not limited just to studying the SMVT. It could be used in a wide range of cellular and molecular research. Conclusion Obviously, SMVT plays an integral role within the human, or drosophila, cell. This protein can be targeted for drug delivery by enhancing the permeability of otherwise poorly-permeable drugs. Research into the use of the SMVT can be led using model systems, such as the drosophila system investigated in this experiment. The homology between drosophila and human genes shows that drosophila could be used as a model system for studying human disease from a genomic standpoint. Works Cited 1. “cDNA Isolation and Analysis: Identifying potential Drosophila melanogaster models of human disease.” Edited by Nelson, K. and Burpee, D. Department of Biology, The Pennsylvania State University, University Park, PA. (2013) 2. Luo S, Kansara VS, Zhu X, et al. “Functional characterization of sodium-dependent multivitamin transporter in MDCK-MDR1 cells and its utilization as a target for drug delivery”. Mol Pharm. 2006;3:329–339. 3. Patel, Mitesh (10/2012). "Molecular expression and functional activity of sodium dependent multivitamin transporter in human prostate cancer cells". International journal of pharmaceutics (0378-5173), 436 (1-2), p. 324. 4. Vadlapudi, Aswani Dutt (01/2013). "Biotin uptake by T47D breast cancer cells: Functional and molecular evidence of sodium-dependent multivitamin transporter (SMVT)". International journal of pharmaceutics (0378-5173), 441 (1-2), p. 535.