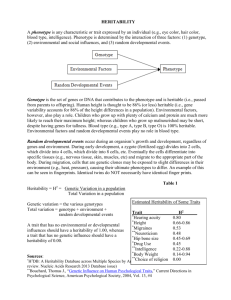
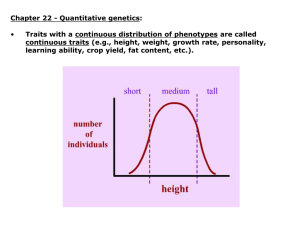
Lecture 13 Selection on quantitative characters
advertisement

Lecture 13 Selection on quantitative characters Selection on quantitative characters What is a quantitative (continuous) character? Selection on quantitative characters What is a quantitative character? • quantitative characters exhibit continuous variation among individuals. Selection on quantitative characters What is a quantitative character? • quantitative characters exhibit continuous variation among individuals. • unlike discrete characters, it is not possible to assign phenotypes to discrete groups. Examples of discrete characters Example of a continuous character Height Two characteristics of quantitative traits: Two characteristics of quantitative traits: 1. Controlled by many genetic loci Two characteristics of quantitative traits: 1. Controlled by many genetic loci 2. Exhibit variation due to both genetic and environmental effects Two characteristics of quantitative traits: 1. Controlled by many genetic loci 2. Exhibit variation due to both genetic and environmental effects • the genes that influence quantitative traits are now called quantitative trait loci or QTLs. Quantitative characters can be controlled by small numbers of genes What are QTLs? What are QTLs? • QTLs possess possess multiple alleles, exhibit varying degrees of dominance, and experience selection and drift. What are QTLs? • QTLs possess multiple alleles, exhibit varying degrees of dominance, and experience selection and drift. • some QTLs exhibit stronger effects than others – these are called major effect and minor effect genes, respectively. What are QTLs? • QTLs possess multiple alleles, exhibit varying degrees of dominance, and experience selection and drift. • some QTLs exhibit stronger effects than others – these are called major effect and minor effect genes, respectively. • the number and relative contributions of major effect and minor effect genes underlies the genetic architecture of the trait. What are QTLs? • QTLs possess multiple alleles, exhibit varying degrees of dominance, and experience selection and drift. • some QTLs exhibit stronger effects than others – these are called major effect and minor effect genes, respectively. • the number and relative contributions of major effect and minor effect genes underlies the genetic architecture of the trait. • mapping QTLs is expensive, labor intensive, and fraught with statistical problems! Mimulus phylogeny F2 progeny from Mimulus cardinalis x M. lewisii F1 hybrids Heritability - heritability does not mean “heritable” or “inherited”!! - heritability represents the degree to which the trait is determined by genetic and not by environmental effects. Heritability Beans: Average is 404 mg. Select the top 10% of the population for next years crop (new mean 692 mg). - the mean of the crop from the selected group is 609 mg. - the average seed size has thus increased by 51% over one generation. 1. What would have occurred if the variation in bean size was entirely due to environmental effects? the mean bean size would have remained unchanged (at about 404 mg). 2. What if all of the variation was controlled by genetic factors? the mean bean size in generation 1 would have been about 692 mg. Heritability Selection differential, S = the “strength” of selection = mean (selected) - mean (whole pop.) = 692 - 404 = 288 Response differential, R = the change in average phenotype due to selection = mean (whole pop. in gen. 1) - mean (whole pop. in gen. 0) = 609 - 404 = 205 Realized heritability, h^2 = R/S = 205/288 = 0.71 - a heritability of 0.71 means that 71% of the variation in bean size in the starting population was due to genetic factors and 29% was caused by the environmental factors Heritability - knowing the heritability of a trait allows us to predict its response to selection. - (Realized heritability, h^2 = R/S) - the equation above can be rearranged to: Response differential, R = h^2 . S - this means that the response of the trait to selection is determined by its heritability and by the intensity of selection. - strong selection acting on a trait with a low heritability will be ineffective! Heritability trait h^2 fingerprint (# of ridges) head width height blood pressure IQ twinning handedness body weight 0.98 0.95 0.84 0.70 0.55 0.52 0.32 0.05 What is heritability? What is heritability? • heritability is the proportion of the total phenotypic variation controlled by genetic rather than environmental factors. What is heritability? • heritability is the proportion of the total phenotypic variation controlled by genetic rather than environmental factors. The total phenotypic variance may be decomposed: VP = total phenotypic variance The total phenotypic variance may be decomposed: VP = total phenotypic variance VG = total genetic variance The total phenotypic variance may be decomposed: VP = total phenotypic variance VG = total genetic variance VE = environmental variance The total phenotypic variance may be decomposed: VP = total phenotypic variance VG = total genetic variance VE = environmental variance VP = VG + VE The total phenotypic variance may be decomposed: VP = total phenotypic variance VG = total genetic variance VE = environmental variance heritability = VG/VP (broad-sense) The total genetic variance (VG) may be decomposed: The total genetic variance (VG) may be decomposed: VA = additive genetic variance The total genetic variance (VG) may be decomposed: VA = additive genetic variance VD = dominance genetic variance The total genetic variance (VG) may be decomposed: VA = additive genetic variance VD = dominance genetic variance VI = epistatic (interactive) genetic variance The total genetic variance (VG) may be decomposed: VA = additive genetic variance VD = dominance genetic variance VI = epistatic (interactive) genetic variance VG = VA + VD + VI The total genetic variance (VG) may be decomposed: VA = additive genetic variance VD = dominance genetic variance VI = epistatic (interactive) genetic variance heritability = h2 = VA/VP (narrow sense) What is additive gene action? What is additive gene action? Consider 2 genes: B1B1 A1A1 A1A2 A2A2 B1B2 B2B2 What is additive gene action? Consider 2 genes: B1B1 B1B2 B2B2 A1A1 0 1 2 A1A2 2 3 4 A2A2 4 5 6 Estimating heritability Estimating heritability • one common approach is to compare phenotypic scores of parents and their offspring: Estimating heritability • one common approach is to compare phenotypic scores of parents and their offspring: Junco tarsus length (cm) Cross Midparent value Offspring value Estimating heritability • one common approach is to compare phenotypic scores of parents and their offspring: Junco tarsus length (cm) Cross F1 x M1 Midparent value 4.34 Offspring value 4.73 Estimating heritability • one common approach is to compare phenotypic scores of parents and their offspring: Junco tarsus length (cm) Cross Midparent value Offspring value F1 x M1 4.34 4.73 F2 x M2 5.56 5.31 Estimating heritability • one common approach is to compare phenotypic scores of parents and their offspring: Junco tarsus length (cm) Cross Midparent value Offspring value F1 x M1 4.34 4.73 F2 x M2 5.56 5.31 F3 x M3 3.88 4.02 Regress offspring value on midparent value ← Slope = h2 Heritability estimates from other regression analyses Comparison Slope Heritability estimates from other regression analyses Comparison Midparent-offspring Slope h2 Heritability estimates from other regression analyses Comparison Slope Midparent-offspring Parent-offspring h2 1/2h2 Heritability estimates from other regression analyses Comparison Slope Midparent-offspring Parent-offspring Half-sibs h2 1/2h2 1/4h2 Heritability estimates from other regression analyses Comparison Slope Midparent-offspring Parent-offspring Half-sibs First cousins h2 1/2h2 1/4h2 1/8h2 Heritability estimates from other regression analyses Comparison Slope Midparent-offspring Parent-offspring Half-sibs First cousins h2 1/2h2 1/4h2 1/8h2 • as the groups become less related, the precision of the h2 estimate is reduced. Heritabilities vary between 0 and 1 Heritability estimates from other regression analyses Comparison Slope Midparent-offspring Parent-offspring Half-sibs First cousins h2 1/2h2 1/4h2 1/8h2 • as the groups become less related, the precision of the h2 estimate is reduced. Cross-fostering is a common approach Q: Why is knowing heritability important? Q: Why is knowing heritability important? A: Because it allows us to predict a trait’s response to selection Q: Why is knowing heritability important? A: Because it allows us to predict a trait’s response to selection Let S = selection differential Predicting the response to selection Example: the large ground finch, Geospiza magnirostris Predicting the response to selection Example: the large ground finch, Geospiza magnirostris Mean beak depth of survivors = 10.11 mm Predicting the response to selection Example: the large ground finch, Geospiza magnirostris Mean beak depth of survivors = 10.11 mm Mean beak depth of initial pop = 8.82 mm Predicting the response to selection Example: the large ground finch, Geospiza magnirostris Mean beak depth of survivors = 10.11 mm Mean beak depth of initial pop = 8.82 mm S = 10.11 – 8.82 = 1.29 mm Q: Why is knowing heritability important? A: Because it allows us to predict a trait’s response to selection Let S = selection differential Let h2 = heritability Q: Why is knowing heritability important? A: Because it allows us to predict a trait’s response to selection Let S = selection differential Let h2 = heritability Let R = response to selection Q: Why is knowing heritability important? A: Because it allows us to predict a trait’s response to selection Let S = selection differential Let h2 = heritability Let R = response to selection R = h2S Predicting the response to selection Example: the large ground finch, Geospiza magnirostris Predicting the response to selection Example: the large ground finch, Geospiza magnirostris Mean beak depth of survivors = 10.11 mm Predicting the response to selection Example: the large ground finch, Geospiza magnirostris Mean beak depth of survivors = 10.11 mm Mean beak depth of initial pop = 8.82 mm Predicting the response to selection Example: the large ground finch, Geospiza magnirostris Mean beak depth of survivors = 10.11 mm Mean beak depth of initial pop = 8.82 mm S = 10.11 – 8.82 = 1.29 mm Predicting the response to selection Example: the large ground finch, Geospiza magnirostris Mean beak depth of survivors = 10.11 mm Mean beak depth of initial pop = 8.82 mm S = 10.11 – 8.82 = 1.29 mm h2 = 0.72 Predicting the response to selection Example: the large ground finch, Geospiza magnirostris Mean beak depth of survivors = 10.11 mm Mean beak depth of initial pop = 8.82 mm S = 10.11 – 8.82 = 1.29 mm h2 = 0.72 R = h2S = (1.29)(0.72) = 0.93 mm Predicting the response to selection Example: the large ground finch, Geospiza magnirostris Mean beak depth of survivors = 10.11 mm Mean beak depth of initial pop = 8.82 mm S = 10.11 – 8.82 = 1.29 h2 = 0.72 R = h2S = (1.29)(0.72) = 0.93 Beak depth next generation = 8.82 + 0.93 = 9.75 mm The selective differential and the selective gradient The selective differential and the selective gradient The selective differential and the selective gradient Selective gradient = β = Selective differential (S) Variance What are heritability estimates in nature? What are heritability estimates in nature? Character Body weight Wing length Tarsus length Bill length Bill depth Bill width Medium Ground finch 0.91 0.84 0.71 0.65 0.79 0.90 • data from Boag (1983) What are heritability estimates in nature? Character Body weight Wing length Tarsus length Bill length Bill depth Bill width Medium Ground finch 0.91 0.84 0.71 0.65 0.79 0.90 Song Sparrow 0.04 0.13 0.32 0.33 0.51 0.50 • data from Boag (1983) and Smith & Zach (1979) What are heritability estimates in nature? What are heritability estimates in nature? Trait Sample size Mean h2 Std. Error What are heritability estimates in nature? Trait Life history Sample size 341 Mean h2 0.262 Std. error 0.012 What are heritability estimates in nature? Trait Sample size Mean h2 Std. error Life history 341 0.262 0.012 Physiological 104 0.330 0.027 What are heritability estimates in nature? Trait Sample size Mean h2 Std. error Life history 341 0.262 0.012 Physiological 104 0.330 0.027 Behavioral 105 0.302 0.023 What are heritability estimates in nature? Trait Sample size Mean h2 Std. error Life history 341 0.262 0.012 Physiological 104 0.330 0.027 Behavioral 105 0.302 0.023 Morphological 570 0.461 0.004 • data from Mousseau and Roff (1983) Natural selection at the phenotypic level 1. Directional selection 2. Stabilizing selection 3. Disruptive selection Natural selection at the phenotypic level 1. Directional selection a form of selection favoring individuals at above or below the mean. - this type of selection causes the trait to either increase or decrease in magnitude and, as a result, reduces the population variance. - example: cranial capacity in early hominid evolution. Directional selection changes the average value of a trait. Normal distribution During selection Number of individuals Before selection After selection Value of a trait For example, directional selection caused overall body size to increase in a cliff swallow population 40 35 30 25 Percentage of birds 20 Nonsurvivors N = 1853 15 10 5 Difference in average 0 40 Survivors N = 1027 35 30 25 20 15 10 5 0 1 2 3 4 5 6 7 8 Body size class 9 10 11 12 Natural selection at the phenotypic level 2. Stabilizing selection a form of selection favoring intermediate phenotypes. - this form of selection reduces variation but does not change the trait’s mean. - example: birth weight in humans. Stabilizing selection reduces the amount of variation in a trait. Normal distribution During selection Number of individuals Before selection High fitness After selection Value of a trait For example, very small and very large babies are most likely to die, leaving a narrower distribution of birthweights. 100 20 50 Mortality 15 30 20 10 Heavy mortality on extremes 10 7 5 5 3 2 0 1 2 3 4 5 7 6 8 Birthweight (pounds) 9 10 11 Percentage of mortality Percentage of Population 70 Natural selection at the phenotypic level 3. Disruptive selection a form of selection favoring both extremes of the phenotypic distribution. - this causes the variation of the trait to increase in the population. - example: beak length in African seedcracker finches. Disruptive selection increases the amount of variation in a trait. Normal distribution During selection Number of individuals Before selection Low fitness After selection Value of a trait For example, only juvenile blackbellied seedcrackers with very long or very short beaks survived long enough to breed. Number of individuals 30 20 10 0 6 7 8 Beak length (mm) 9 10 11 Selection on quantitative traits - the three forms of selection outlined above occur on what are called quantitative or polygenic traits. - quantitative traits differ from discrete traits in that it is not possible to assign individuals into distinct classes. Selection on quantitative traits 1. vary in a continuous fashion among individuals 2. are controlled by many genetic loci. 3. are affected by both genetic and environmental factors. - to understand and predict the evolution of quantitative characters, we must define an important parameter called heritability. Modes of selection on quantitative traits Modes of selection on quantitative traits Directional selection on oil content in corn Modes of selection on quantitative traits Stabilizing selection on gall size Modes of selection on quantitative traits Disruptive selection in black-bellied seedcracker finches