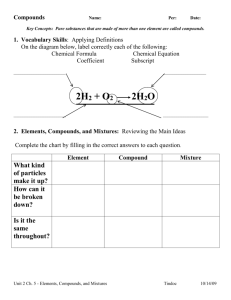
American Psychological Association 5th Edition
advertisement

IN SILICO TARGET PREDICTION BY TRAINING NAÏVE BAYESIAN MODELS ON CHEMOGENOMICS DATABASES Nidhi Submitted to the faculty of the Chemical Informatics Graduate Program in partial fulfillment of the requirements for the degree Master of Science in the School of Informatics, Indiana University December 2005 Accepted by the Faculty of Indiana University, in partial fulfillment of the requirements for the degree of Master of Science ________________________ Mahesh Merchant, Ph.D., Chair _______________________ Jeremy Jenkins, Ph.D. _______________________ Douglas Perry, Ph.D. ii ACKNOWLEDGEMENTS I extend my gratitude and appreciation to the people who made this master’s thesis possible. My foremost thanks go to my research advisor, Dr. Jeremy Jenkins. I want to thank him for his insights and suggestions that helped me have a deeper understanding of the subject matter. I am grateful to Dr. Mahesh Merchant for the support and encouragement that I received from him throughout this project. I am indebted to Dr. Douglas Perry for his invaluable guidance and support right from the very beginning of my work. I would also like to acknowledge my manager Dr. John Davies and colleague Dr. Meir Glick at Novartis Institutes for Biomedical Research for their constant feedback on my work which made me strive to do better. Many thanks go to the faculty and staff at School of Informatics. These include Dr. Kelsey Forsythe, Dr. David Wild, Ms. Mary O’Neil, Ms.Vickie Bucker and Mr. Dale Ray. Special thanks are due to MDL for awarding me with the Elsevier MDL Excellence in Informatics fellowship. Last but not the least, I thank my parents and my brother for always being there for me and supporting me in all my endeavors. iii TABLE OF CONTENTS ACKNOWLEDGEMENTS ............................................................................................... iii LIST OF TABLES .............................................................................................................. v LIST OF FIGURES ........................................................................................................... vi INTRODUCTION .............................................................................................................. 1 METHODS ......................................................................................................................... 9 RESULTS ......................................................................................................................... 18 DISCUSSION ................................................................................................................... 32 CONCLUSION ................................................................................................................. 35 REFERENCES ................................................................................................................. 36 APPENDIX ....................................................................................................................... 39 iv LIST OF TABLES Table 1. Datasets used for training and testing the naïve Bayesian models Table 2. MDDR activity classes used for study Table 3. Murcko assemblies for COX-2 inhibitors derived from WOMBAT database Table 4. Biggest target predicted by naïve Bayes model for MDDR generic activity classes v LIST OF FIGURES Figure 1a. Chemogenomics approach to drug discovery Figure 1b. Organization of a Chemogenomics Database Figure 2a. Substructure search vs Extended connectivity fingerprint structure search Figure 2b. Extended Connectivity Fingerprint generation method Figure 3. Pipeline Pilot protocol for building multiclass naïve Bayes model Figure 4. Murcko assembly distribution of MDDR and WOMBAT. Figure 5a-b. Percentage of test compounds with order of prediction for WOMBAT85% model Figure 6a. Percentage of test compounds derived from ten MDDR activity classes with order of prediction for WOMBAT model Figure 7a. Top target distribution for MDDR activity class Antineoplastic Figure 7b. Top target distribution for MDDR Kinases Figure 7c. Top target distribution for MDDR activity class Antiinflammatroy, Intestinal Figure 7d. Top target distribution for MDDR activity class Antihypertensive Figure 7e. Top target distribution for MDDR activity class Antiarithritic Figure 8a. MDDR Extreg: 142057 (N-(2-aminophenyl)-4-(3-hydroxypropanamido) benzamide). A substructure in many compounds with anti-cancer activity and predicted as a HDAC inhibitor. Figure 8b. MDDR Extreg 142055 (N-(2-aminophenyl)-4-isobutyramidobenzmide). Analog of 142057 and predicted as a HDAC inhibitor Figure 8c. N-aryl benzamide class that is patented as a HDAC inhibitor class Figure 9a. Trichostatin D. MDDR Extreg: 286017. Predicted as a HDAC inhibitor vi Figure 9b. Trichostatin A. Known HDAC inhibitor Figure 10. Closest similarity histogram for MDDR and WOMBAT COX-1 inhibitors Figure 11. Tanimoto similarity coefficient vs. the order of prediction for MDDR Kinases vii INTRODUCTION The completion of Human Genome Project is seen as a gateway to the discovery of novel drug targets (Jacoby, Schuffenhauer, & Floersheim, 2003). How much of this information is actually translated into knowledge, e.g., the discovery of novel drug targets, is yet to be seen. The traditional route of drug discovery has been from target to compound. Conventional research techniques are focused around studying animal and cellular models which is followed by the development of a chemical concept. Modern approaches that have evolved as a result of progress in molecular biology and genomics start out with molecular targets which usually originate from the discovery of a new gene .Subsequent target validation to establish suitability as a drug target is followed by high throughput screening assays in order to identify new active chemical entities (Hofbauer, 1997). In contrast, chemogenomics takes the opposite approach to drug discovery (Jacoby, Schuffenhauer, & Floersheim, 2003). It puts to the forefront chemical entities as probes to study their effects on biological targets and then links these effects to the genetic pathways of these targets (Figure 1a). The goal of chemogenomics is to rapidly identify new drug molecules and drug targets by establishing chemical and biological connections. Just as classical genetic experiments are classified into forward and reverse, experimental chemogenomics methods can be distinguished as forward and reverse depending on the direction of investigative process i.e. from phenotype to target or from target to phenotype respectively (Jacoby, Schuffenhauer, & Floersheim, 2003). The identification and characterization of protein targets are critical bottlenecks in forward 1 Homologous Proteins Gene Families Genomics/ Proteomics Drug Chemotypes Chemoproteomics Figure 1a. Homologous proteins are characterized by studying gene families and traditional drug discovery route is from protein targets to drug molecules. Chemogenomics explores the relationship between structure (“chemical genotype”) and biological activity (“chemical phenotype”) and establish genetic connection based on chemical phenotype. Protein1 Compound1 Compound3 S i m i l a r i t y Cheminformatics Bioinformatics Protein2 Compound2 Figure 1b.Cheminformatics techniques are based on structural similarity between molecules while bioinformatics deals with sequence similarity to establish homologous proteins. Chemogenomics can be used to derive the missing links between compounds and targets as well as genetic connections between two targets. 2 chemogenomics experiments. Currently, methods such as affinity matrix purification (Taunton, Hassig, & Schreiber, 1996) and phage display (Sche, McKenzie, White, & Austin, 1999) are used to determine targets for compounds. None of the current techniques used for target identification after the initial screening are efficient. In silico methods can provide complementary and efficient ways to predict targets by using chemogenomics databases to obtain information about chemical structures and target activities of compounds. Annotated chemogenomics databases integrate chemical and biological domains and can provide a powerful tool to predict and validate new targets for compounds with unknown effects (Figure 1b). A chemogenomics database contains both chemical properties and biological activities associated with a compound. The MDL Drug Data Report (MDDR) (Molecular Design Ltd., San Leandro, California) is one of the well known and widely used databases that contains chemical structures and corresponding biological activities of drug like compounds. The relevance and quality of information that can be derived from these databases depends on their annotation schemes as well as the methods that are used for mining this data. In recent years chemists and biologist have used such databases to carry out similarity searches and lookup biological activities for compounds that are similar to the probe molecules for a given assay. With the emergence of new chemogenomics databases that follow a wellstructured and consistent annotation scheme, new automated target prediction methods are possible that can give insights to the biological world based on structural similarity between compounds. The usefulness of such databases lies not only in predicting targets, but also in establishing the genetic connections of the targets discovered, as a consequence of the prediction. 3 The ability to perform automated target prediction relies heavily on a synergy of very recent technologies, which includes: i) Highly structured and consistently annotated chemogenomics databases. Many such databases have surfaced very recently; WOMBAT (Sunset Molecular Discovery LLC, Santa Fe, New Mexico), KinaseChemBioBase (Jubilant Biosys Ltd., Bangalore, India) and StARLITe (Inpharmatica Ltd., London, UK), to name a few. ii) Chemical descriptors (Xue & Bajorath, 2000) that capture the structure-activity relationship of the molecules as well as computational techniques (Kitchen, Stahura, & Bajorath, 2004) that are specifically tailored to extract information from these descriptors. iii) Data pipelining environments that are fast, integrate multiple computational steps, and support large datasets. A combination of all these technologies may be employed to bridge the gap between chemical and biological domains which remains a challenge in the pharmaceutical industry. 4 BACKGROUND The current trend in drug discovery is to focus on gene families, which makes it possible to work with multiple targets simultaneously, because of their genetic interrelationship. Chemogenomics seeks to apply this relationship by starting with chemical compounds to find novel targets. In silico methods can be used to further advance this field. A number of different in silico approaches are possible, depending on the objective of a particular study. The most widely used method to make a selection is to carry out similarity searches on a structural compound database with reference molecules (Hert et al., 2004). Cutoff values derived from similarity metrics like the Tanimoto coefficient (Willett, 2005) can be used to filter uninteresting compounds. Three-dimensional pharmacophorebased searches can also be used when 3-D structural information and active ligands are available (Toledo, Lydon, & Elbaum, 1999). Classification methods are useful to generate focused libraries as well as identify novel compounds from large compound collections. Genetic algorithms (GA) (Gillet, Willett, & Bradshaw, 1998), neural networks (NN) (Manallack et al., 2002), support vector machines (SVM) (Weinstein et al., 1997), recursive partitioning (RP) (P. Blower, Fligner, Verducci, & Bjoraker, 2002; Rusinko, Farmen, Lambert, Brown, & Young, 1999), naïve Bayes classifier (NB) (Klon, Glick, & Davies, 2004; Xia, Maliski, Gallant, & Rogers, 2004), and standard and partial least square methods (Zhu, Hou, Chen, & Xu, 2001) are some of the classification techniques that have gained popularity among researchers to model structure-activity relationships of compounds. Clustering methods are also widely used to group compounds with similar bioactivity based on their structural information (Shemetulskis, Dunbar, Dunbar, 5 Moreland, & Humblet, 1995). A number of hierarchical methods such as Wards and group-average and non hierarchical clustering methods such as Jarvis Patrick were compared in a study conducted by Brown and Martin (Brown & Martin, 1998). The Wards clustering method is reported to perform best in separating active from inactive compounds in this study. K-means is another nonhierarchical clustering method that has been reported to perform well in a study to identify ligand binding sites on proteins (Glick, Robinson, Grant, & Richards, 2002). The performance of a particular similarity search, clustering or classification method in terms of predicting activity for a given compound depends on the dataset as well the choice of descriptors. Numerous descriptors have been developed to describe molecules: 1-D descriptors such as molecular weight, Simplified Molecular Input Line Entry Specification (SMILES), and melting point, 2-D descriptors such as connectivity tables and 2-D fingerprints, and 3-D descriptors such as pharmacophore definition triplets (PDTs) (Abrahamian et al., 2003) have been used to represent chemical structures. 2-D fingerprinting is by far the most commonly used structure abstraction technique. A recent study done by Hert et al. to compare a number of 2-D fingerprints such as BCI, Daylight, Extended Connectivity fingerprints (ECFPs), Unity and Avalon shows the effectiveness of ECFPs, which are based on circular substructures, over other topological descriptors (Hert et al., 2004). Also, Klon et al. have reported considerable enrichment in high throughput docking data by using ECFPs in combination with Naïve Bayesian classifier (Klon, Glick, & Davies, 2004). Modeling biological activities based on chemical structure abstraction for a wide range of compounds has been attempted recently in the form of a computer program, 6 PASS (Lagunin, Stepanchikova, Filimonov, & Poroikov, 2000) that has also been integrated with the National Cancer Institute’s open database browser (Poroikov et al., 2003). PASS makes use of substructure descriptors called multilevel neighborhoods of atoms (MNA) (Fomenko, Sobolev, Filimonov, & Poroikov, 2003). After an initial training with known biologically active compounds, it can be used to predict a wide range of biological activities such as mutagenicity, carcinogenicity, and embryotoxicity for test compounds. The usefulness of annotated chemical libraries, overlaying the ligand-target domains, has been recently recognized. Studies analyzing ligand-target space (Weinstein et al., 1997) and finding correlation between substructures and gene expression patterns (P. E. Blower et al., 2002) have been reported. Kohonen self-organizing maps were recently employed in an attempt to classify 20,000 compounds tested against 80 of NCI’s tumor cell lines (Rabow, Shoemaker, Sausville, & Covell, 2002) with very useful results. In another effort to integrate bio- and cheminformatics domains, annotation schemes for ligands of four target classes (enzymes, G protein-coupled receptors, nuclear receptors and ligand-gated ion channels) have been proposed (Schuffenhauer et al., 2002). The amalgamation of a robust fingerprinting method and an appropriate computational model trained on a well-annotated chemogenomics database can be an extremely useful tool to discover novel targets for compounds with unknown activities. Further, the structure-activity relationship homology principle (Frye, 1999) can be applied to discover leads for homologous targets. 7 Hypothesis It is possible to predict targets for compounds with unknown activities by training multiple naïve Bayes classifiers on compounds with known biological targets. The intent of this research is to train multiple Laplacian-modified naïve Bayes models with extended connectivity fingerprints on compound records derived from the WOMBAT database in order to predict targets for test compounds derived from the MDDR database. 8 METHODS Extended Connectivity Fingerprints Extended connectivity fingerprints, developed by SciTegic (SciTegic, San Diego, California), are structure fingerprints based on the Morgan algorithm (Morgan, 1965). An ECFP feature represents an exact structure with limited and specified attachment points. Therefore, a substructure search using the fragment para-substituted benzene (a) would return compounds (b, c) while a search with ECFPs would return only (c) (Figure 2a). This is because for ECFPs, (b) does not conatin (a) as there is substitution on the benzene ring at locations other than the specified attachment atoms marked as X. a b c Figure 2a Substructure search vs Extended connectivity fingerprint structure search ECFPs are generated in an iterative fashion. Initially, each atom is assigned a code that is derived from the number of connections to the atom, element type, charge and mass of the atom which corresponds to ECFP with a neighborhood size of “0”. In the first iteration, information about each atom’s immediate neighbors is collected and a new code representing the atom and its immediate neighbors is generated. In each iteration, the neighborhood size becomes larger and the updated codes of the atoms from previous iterations are used for assigning new codes. When the desired neighborhood size is reached, the set of all features is returned as a fingerprint (Figure 2b). 9 Figure 2b Extended connectivity fingerprint generation method ECFPs with a neighborhood size of 6 (i.e., ECFP_6) were used as structural descriptors to train the naïve Bayes classifiers as they represent a fairly large set of features or structural units that are crucial for structural comparison of compounds (Klon, Glick, Thoma, Acklin, & Davies, 2004). Multiclass Naïve Bayesian Modeling Naïve Bayes Naïve Bayes, a statistical classification method, can be efficiently used for large datasets as it scales linearly with the number of molecules. It is based on Bayes rule of conditional probability (Weisstein) which states that given two events A and B, the probability of occurrence of event A given that event B has already occurred, P(A|B), is given by: P(A|B) =P(B|A)P(A)/P(B) where P(A) and P(B) are the probability of occurrence of events A and event B, respectively. In this study, naïve Bayes is used to classify compounds into actives and inactives against a particular target based on the frequency of occurrence of a given feature, which is encoded by extended connectivity fingerprint bits. Given an ECFP, the probability of a 10 given compound being active against a particular target that contains ECFP, is given by P(active|ECFP), where event active refers to the event that the compound is active. Thus, P(active|ECFP) = P(ECFP|active) P(active)/P(ECFP) This method is said to be naïve because it naively assumes independence of the features from one another. Making this assumption, it is valid to multiply the individual feature probabilities. Given n features or ECFP bits, P(active|ECFP) = P(ECFP1|active) X P(ECFP2|active) X P(ECFPn|active)P(active)/P(ECFP) Laplacian Correction A Laplacian correction is used as different features are sampled at different frequencies. If N is the number of compounds in a given dataset, of which M are active, the baseline probability of a compound being active is given by: P(active) = M/N The probability of a compound being active given feature F present in B number of compounds of which A are active can be given by P(active|F) = A/B As the number of compounds B diminishes, the probability estimate of a feature starts getting biased. For example if A =1 and B =1 then P(active|F) = 1, which is not true as the feature is undersampled. 11 To correct for the above anomaly, it is assumed that most of the features have no relationship with activity, i.e., for most Fi, P(active|Fi) = P(active), which is the baseline probability. If the feature is sampled X additional times, then P(active) X X of those new samples should be active. Therefore, each feature can be corrected by adding those virtual samples. P(active|F) = (A + P(active) X X) / (B + X) Now, as the number of samples, B, decreases, the feature’s probability contribution converges to P(active). Relative Estimator Naïve Bayes as implemented in Pipeline Pilot is a relative estimator which is obtained by dividing the Laplacian-corrected Bayes classifier by P(active). Thus, P(active|F) = P(active|F)/P(active) which implies: i) for most features, log P(active|F) ~ 0 ii) for features more common in actives, log P(active|F) > 0 iii) for features less common in actives, log P(active|F) < 0. The Learn Molecular Categories component in Scitegic’s Pipeline Pilot software can build multiple naïve Bayes models in one protocol. The user needs to specify a “CategoryProperty” property on each data record, in this case target name, which is subsequently used to create different equations for different values of that property. Once a multiple naïve Bayes model is created, it can be added to another protocol to calculate naïve Bayes scores for multiple targets. A test compound is passed through each of the equations of this model and scores corresponding to each target name is calculated. The 12 highest score for a target is taken as most likely target for that compound. Similarly, the next highest score for a target can be assigned as second most likely target and so on. Databases and Datasets WOMBAT WOMBAT (World of Molecular Bioactivity) is a consistently annotated chemogenomics database. The version used for this study, WOMBAT 2005.1, contains 117,007 compounds and 104,230 unique SMILES. 230,000 biological activities and 1,021 unique targets are reported for these compounds. It contains 4,786 different chemical series from 4,773 papers published in medicinal chemistry journals between 1975 and 2004. The database follows a hierarchical scheme such that it is possible to look up the target family. The target families are based on the functional properties of targets. Targets are usually proteins but can also be DNA or RNA. For the purpose of this study, compounds with activity threshold, IC50/EC50/Ki/Kb/Kd/MIC/ED50 < 30µm were selected. Thus, a reduced dataset of 103,735 compounds with 964 biological targets was generated. Also, WOMBAT is organized in a way in which one compound may have more than one target. The database was preprocessed to associate one target per compound so that there is unique value for the “CategoryProperty” for the Learn molecular component of Pipeline Pilot. As a result, 192,373 compound records were generated because of duplication. This dataset will be referred to henceforth as the WOMBAT database. MDDR MDDR contains information about biologically relevant compounds. Many biological activities reported in MDDR are very generic e.g. antineoplastic, antihypertensive and 13 anti-inflammatory. It covers information available in patent literature, journal articles, and meetings. MDDR is updated annually. The version used for this study contains 159,967 compounds and 761 biological activity classes. There are 2,789 compounds in MDDR with no structural information. Since the descriptors used for statistical analysis in this study are based on structural information, these compounds were removed from the dataset and the remaining 156,873 compounds with 659 biological activity classes were kept for the study. Datasets We built two separate multiclass Bayes models for the purpose of this study. The WOMBAT 85% Model has 85% of WOMBAT compound records as training set and the other 15% is used as the test set. Cross validation of WOMBAT database was carried out because there are large number of records available for the study. There is no duplication of compounds across two sets. The second model, the WOMBAT model, uses WOMBAT as the training set. Test sets are derived from ten MDDR specific biological activity classes and ten generic biological activity classes. Table 1 summarizes the number of compounds and respective databases from which training and test sets are extracted for both models. Table 2 shows the MDDR activity classes with the total number of compound records corresponding to each class. Model Name Wombat85% Model Wombat Model Number of compounds Training Set Test Set 162238 30135 (WOMBAT) (WOMBAT) 192373 53137 (WOMBAT) (MDDR) Table 1. Datasets used for training and testing the naïve Bayesian models 14 MDDR Activity Class Number of Compounds Phosphodiesterase IV Inhibitor 2000 Cyclooxygenase-1 Inhibitor 88 Cyclooxygenase-2 Inhibitor 1055 Acetylcholinesterase Inhibitor 810 Angiotensin II AT1 Antagonist 2185 Angiotensin II AT2 Antagonist 53 ACE Inhibitor 570 Reverse Transcriptase Inhibitor 819 HIV-1 Protease Inhibitor 1027 H+/K+-ATPase Inhibitor 751 Antineoplastic 19621 Kinases* 1858 Anti-inflammatory, Intestinal 29 Antihypertensive 11124 Anti-arthritic 11147 Table 2. MDDR activity classes used for study *Kinases are derived from six MDDR activity classes viz. Adenosine Kinase Inhibitor, Nucleoside Diphosphate Kinase Inhibitor, Phosphatidylinositol Kinase Inhibitor, Protein Kinase C Inhibitor, Thymidine Kinase Inhibitor and Tyrosine-Specific Protein Kinase Inhibitor Software Scitegic’s Pipeline Pilot™ version 4.5.2 was employed to build the multiclass naïve Bayesian models. Hardware The computations were carried out on a Pipeline Pilot server with Intel® Xeon™ 2.7 GHz dual processors and 4 GB of RAM. 15 Protocol for Building Multiclass Naïve Bayesian Models Figure 3 shows the components used by the Pipeline Pilot protocol that was used to build multiclass Bayesian models. The preprocessing steps involved: a. Reading of the compound library into the protocol b. Conversion to Pipeline Pilot molecule format from SMILES string c. Removal of salts if any from the molecule d. Standardization of stereo-atoms and charges e. Reformatting of text string contained in target name to remove any occurrences of single quotes f. Calculation of extended connectivity fingerprints for all molecules Figure 3 Pipeline Pilot protocol for building multiclass naïve Bayes models The protocol generates a new component called Learn Molecular properties. This component can be used in another protocol with test data as input to this component. Diversity Analysis Diversity analysis is carried out to ensure that the model is applicable over a wide range of structural classes. An analysis of structural diversity among the training and test set can thus establish the effectiveness of the computational model. The chemical diversity was assessed by carrying out Murcko assemblies analysis (Bemis & Murcko, 1996) of WOMBAT and MDDR compounds. Murcko assemblies are obtained by removing the side chains from the molecule and keeping the core ring system and associated atom type, 16 bond order and hybridization information. Table 3 shows examples of COX-2 inhibitors derived from the WOMBAT database, and their corresponding Murcko assemblies. After generating Murcko assemblies for all the compounds in WOMBAT and MDDR, unique as well as assemblies common to both can be determined. Based on the number of unique assemblies it can be determined if two datasets differ significantly in terms of chemical diversity. Compound Structure Murcko Assembly Table 3. Murcko assemblies for COX-2 inhibitors derived from WOMBAT database. 17 RESULTS Diversity Analysis Figure 4 shows the results of the diversity analysis based on Murcko scaffold analysis. It shows that there are 65,806 Murcko assemblies unique to MDDR, 24,772 assemblies unique to WOMBAT and 6,395 assemblies common to both datasets. Also, the figure depicts the frequency of assemblies in the datasets, e.g., the bar region corresponding to blue accounts for Murcko assemblies that occur just once in the complete dataset (singletons). The red region corresponds to assemblies that occur twice in the dataset, and so on. The results show that MDDR is more diverse than WOMBAT, as it contains larger number of distinct Murcko fragments, but it cannot be used for effective target prediction simply because it is not highly annotated. There is a significant overlap between both datasets, but in spite of that there are large numbers of Murcko fragments unique to each database. 18 M- MDDR Scaffolds; MC, WC-Common Scaffolds in MDDR & WOMBAT; W-WOMBAT Scaffolds Frequency of scaffold occurrence denoted by color; blue: 1 occurrence, red: 2 occurrences, yellow: 3 occurrences, black: 4 occurrences, green: 5 or more occurrences Figure 4. Murcko assembly distribution of MDDR and WOMBAT Method Validation Wombat 85% Model The top 3 scores or top 3 targets were evaluated and the results were interpreted in terms of percentage of compounds and rank in which the model predicted the right target. Figure 5a shows the results of target prediction for the WOMBAT 85% model. This model predicts the right target on the first guess for 82% of the compounds, on the second guess for 8% of the compounds, and on the third guess for 2% of the compounds. 19 Also, 8% of the compounds were not correctly predicted in the top 3 guesses. A further analysis was done to examine predicted target families. The model predicts the right target family for 89% of the compounds in first guess as shown in, Figure 5b. First Target Prediction right Second Target prediction right Third Target Prediction right Not predicted right in top three predictions Figure 5a Target Prediction 20 First Target Prediction right Second Target prediction right Third Target Prediction right Not predicted right in top three predictions Figure 5b Target Family Prediction Figure 5a-b. Percentage of test compounds with order of prediction for WOMBAT85% model WOMBAT Model Figure 6a shows the percentage of compounds predicted with the correct target in the top three predictions, as well as compounds for which the model failed to predict the correct target. Figure 6b correspondingly depicts the percentage of compounds that were predicted in the correct target family, as well as the unsuccessful predictions for the target family. On average, 77% of the compounds were predicted with correct targets and 79% of the compounds were predicted with the correct family in top three predictions. 21 First Target Prediction right Second Target prediction right Third Target Prediction right Not predicted right in top three predictions Figure 6a Target Prediction 22 First Target Family Prediction right Second Target Family prediction right Third Target Family Prediction right Not predicted right in top three predictions Figure 6b Target Family Prediction Figure 6a-b Percentage of test compounds derived from ten MDDR activity classes with order of prediction for WOMBAT model Qualitative Analysis Top targets predicted for five generic activity classes, viz. Antineoplastic, Kinase, Antiarthritic, Antihypertensive and Anti-inflammatory, Intestinal, derived from MDDR were examined. The results were binned on target frequencies and interpreted in terms of the type of targets assigned to the generic activities by naïve Bayes Models. Figures 7a-e show the distributions of targets for five generic activity classes. Verification with the 23 scientific literature available on corresponding targets revealed that the predicted target with maximum frequency of compounds was associated with the corresponding generic activity. Antineoplastics have tubulin, a well known antineoplastic target, as the biggest target (8%) (Figure 7a) (Hadfield, Ducki, Hirst, & McGown, 2003). Tubulin is essential to many vital processes such as cell division and movement of materials within a cell. Drugs such as Taxol® bind to tubulin and cause the protein to lose its flexibility, which prevents cell division. Figure 7a. Target Distribution: Top targets predicted for generic MDDR activity class Antineoplastic. Unregulated kinase activity is a frequent cause of cancer where kinases regulate aspects that control cell growth, movement and death (Melnikova & Golden, 2004). 24 Therefore many kinases are anticancer targets, which is evident by the target distribution for Antineoplastics. Kinases have epidermal growth factor receptor (EGFR), which is a tyrosine protein kinase (Mendelsohn & Baselga, 2003), as the biggest target (27%) (Figure 7b). These receptors exist on the cell surface and are involved in initiating a signal transduction cascade which ultimately leads to DNA synthesis and cell proliferation. Figure 7b Target Distribution: Top targets predicted for MDDR Kinases* The Anti-inflammatory, Intestinal activity class has NK1 as the biggest target (Figure 7c). NK1 is a tachykinin receptor. Tachykinin receptor antagonists have been strongly implicated for the treatment of at least 6 major disease groups which includes CNS disorders, pain and emesis, airway disease, urinary incontinence and intestinal dysfunction (Khawaja & Rogers, 1996). 25 Figure 7c Target Distribution: Top targets predicted for MDDR activity class Anti-inflammatory, Intestinal Antihypertensives have Angiotensin II type 1 (AT1) receptor as their biggest target (Figure 7d). AT1 receptor has a well established role in maintaining blood pressure, and water and electrolyte homeostasis (Wexler et al., 1992). AT1 antagonists work as antihypertensive agents by causing either arterial or mixed arterial and venous dilation. 26 Figure 7d Target Distribution: Top targets predicted for MDDR activity class Antihypertensive The Antiarthritic class of compounds has COX-2 as the biggest target (14%). COX-2 is an enzyme responsible for mediating pain and inflammation (Figure 7e). inhibitors are often used to treat arthritis (Hochberg, 2005). 27 COX-2 Figure 7e Target Distribution: Top targets predicted for MDDR activity class Antiarthritic Table 4 summarizes the biggest assigned target for each activity class along with binning frequency and respective the target families. MDDR Activity Class Binning Frequency Biggest Target Target Family Antineoplastic 50 Tubulin Tubulin Kinases* 10 EGFR Tyrosine protein Kinase Anti-inflammatory, Intestinal Antihypertensive Not Binned NK1 GPCR 50 AT1 GPCR Antiarthritic 50 COX-2 Prostaglandin G/H synthase, oxidoreductase, peroxidase, dioxygenase Table 4. Biggest target as predicted by naïve Bayes Models for MDDR generic activity classes 28 Examples of compounds with right prediction Histone Deacytlase (HDAC) inhibitors have been recently recognized as anticancer agents due to their role in the regulation of transcription (Vigushin & Coombes, 2002). HDAC, when inhibited, stops differentiation and induces cell cycle arrest. For nearly 2% of MDDR compounds with antineoplastic activity, HDACs are predicted as top targets by naïve Bayesian Models (Figure 7a) though MDDR contains no information on specific targets for these compounds. I here report three such compounds which have high structural similarity to the known HDAC inhibitors. i) N-(2-aminophenyl)-4-(3-hydroxypropanamido) benzamide (Figure 8a) MDDR Extreg 142057 is predicted as a HDAC inhibitor. This compound is a substructure in several other compounds that have antineoplastic activity. Also, one of these compounds belongs to a class of compounds known as N-aryl benzamides which are synthetic HDAC inhibitors (D. F. Schuppan, 2003). Figure 8c shows the Markush representation of the entire class and example of a compound belonging to that class. ii) N-(2-aminophenyl)-4-isobutyramidobenzmide (Figure 8b) MDDR Extreg 142055 is an analogue of N-(2-aminophenyl)-4-(3-hydroxypropanamido) benzamide and occurs as a substructure in various compounds with anticancer activity. HDAC is predicted as the top target for this compound. 29 Figure 8a N-(2-aminophenyl)-4-(3-hydroxypropanamido) benzamide Figure 8b N-(2-aminophenyl)-4-isobutyramidobenzmide Figure 8c N-aryl benzamide class iii) MDDR Extreg 286017: Trichostatin D (Figure 9a) is listed as an Antineoplastic in MDDR with no reported literature on a specific target. The top target for this compound as predicted by naïve Bayes Models was HDAC. Trichostatin A (Figure 9b) is a known natural HDAC inhibitor (Vigushin et al., 30 2001). It can be seen that both compounds have the same chemotypes except for the extra sugar ring in Trichostatin D. A further discussion with a chemist working on HDAC inhibitors in Novartis revealed that the compound can be very likely a prodrug. A prodrug is a pharmacological compound which is administered in an inactive form and is metabolized into an active form in the body. The sugar ring would be cleaved off in vivo resulting in a hydroxamic acid group as present in Trichostatin A. HO OH H N N O O OH O OH O Figure 9a. Trichostatin D 7-(4-dimethylaminophenyl)-4,6-dimethyl-7-oxo-N-3,4,5-trihydroxy-6- (hydroxymethyl) tetrahydro-2H-pyran-2-yloxy) hepta-2,4-dienamide Figure 9b. Trichostatin A 7-(4-dimethylaminophenyl)-4,6-dimethyl-7-oxo-hepta-2,4-dienehydroxamic acid 31 DISCUSSION Naïve Bayesian modeling in conjunction with extended connected fingerprints has been shown to perform well in identifying actives in comparison to other substructure based searching methods (Klon, Glick, & Davies, 2004). I here investigated the application of the Multicategory Naïve Bayes Model in predicting targets for compounds with unknown or little known activity information. The method is applicable over a wide range of activity classes and 77% of the test compound sets used for validation are predicted with correct targets. The method also performs well while correlating targets with the generic activities. The training of the Multicategory Naïve Bayes Model with a dataset of ~190,000 compound records with 964 targets takes nearly six hours. The subsequent target prediction rate is about 100 minutes for 100,000 compounds. Thus, it can be efficiently used as a complementary tool for target prediction for a big library of compounds. Data mining algorithms are largely dependent on the training datasets. First, the structural diversity among underlying compounds is essential to the success of such algorithms. In this study, COX-1 inhibitors have the lowest correct prediction percentage. Only ~27% of the COX-1 inhibitors are successfully predicted by the algorithm. To account for such a low prediction, I carried out a closest Tanimoto similarity analysis between the test set (COX-1 compound sets derived from MDDR) and training set (COX-1 compound sets derived from WOMBAT). Figure 10 shows the percentage of MDDR compounds with a given Tanimoto Similarity coefficient value with WOMBAT compounds. Nearly 95% of the MDDR compounds have a Tanimoto value <0.4 with WOMBAT COX-1 compounds. 32 Closest Similarities COX-1 % of MDDR Samples 30% 25% 20% 15% Frequency 10% 5% 0% 0 0.2 0.4 0.6 0.8 1 Tanimoto Similarities Figure 10. Closest Tanimoto similarity distribution for MDDR COX-1 compounds This implies that WOMBAT COX-1 compounds do not have sufficient structural diversity. The use of a structurally diverse dataset for training would thus result in better predictions. Second, high similarity between compounds in training and test sets can lead to good predictions. Further analysis of the kinases derived from MDDR was conducted to check how well the Bayes classifier works when there is low similarity between test and training sets. Figure 11 is a plot of Tanimoto similarity vs. the order of prediction for MDDR kinases. For a high similarity between the MDDR and WOMBAT, the first prediction is almost always correct. There is a decrease in correct prediction percentage with decrease in similarity but even for a low similarity score of 0.2-0.5, correct predictions are made by naïve Bayes classifier. This shows that Bayes can classify data even at low similarity values. 33 Not Predicted as Kinase in top 10 predictions Figure 11. Tanimoto similarity coefficient and the order of correct prediction for MDDR kinases Third, the consistency and quality of annotation in chemogenomics database is of prime importance when it comes to deriving meaningful and reliable conclusions from the predictions. The information contained in such databases is derived from research data published in medicinal chemistry journals, patents, and other such sources. There is a constant need for updating such databases as information about new targets and compounds becomes available. 34 CONCLUSION Naive Bayes models trained on a well-annotated chemogenomics database can be used to predict targets for compounds with unknown activities and ascertain genetic connections between targets. Structural diversity among compounds used for building models plays an important role in automated target prediction methods. Therefore, using a structurally diverse chemogenomics database as training set would result in better predictions. The use of in silico techniques in abstracting the structure activity relationship is valid as long as underlying information is correct and up-to-date. As new discoveries are made in the biological and chemical world, new information should be incorporated as it becomes available. 35 REFERENCES Abrahamian, E., Fox, P. C., Naerum, L., Christensen, I. T., Thogersen, H., & Clark, R. D. (2003). Efficient generation, storage, and manipulation of fully flexible pharmacophore multiplets and their use in 3-D similarity searching. J Chem Inf Comput Sci, 43(2), 458-468. Bemis, G. W., & Murcko, M. A. (1996). The properties of known drugs. 1. Molecular frameworks. J Med Chem, 39(15), 2887-2893. Blower, P., Fligner, M., Verducci, J., & Bjoraker, J. (2002). On combining recursive partitioning and simulated annealing to detect groups of biologically active compounds. J Chem Inf Comput Sci, 42(2), 393-404. Blower, P. E., Yang, C., Fligner, M. A., Verducci, J. S., Yu, L., Richman, S., et al. (2002). Pharmacogenomic analysis: correlating molecular substructure classes with microarray gene expression data. Pharmacogenomics J, 2(4), 259-271. Brown, R. D., & Martin, Y. C. (1998). An evaluation of structural descriptors and clustering methods for use in diversity selection. SAR QSAR Environ Res, 8(1-2), 23-39. D. F. Schuppan, C. H., M. Gansmayer, M. Ocker, K. Thierauch. (2003). Germany Patent No. WO 2004058234;US 2005054647. Fomenko, A. E., Sobolev, B. N., Filimonov, D. A., & Poroikov, V. V. (2003). [Use of structural MNA descriptors for designing profiles of protein families]. Biofizika, 48(4), 595-605. Frye, S. V. (1999). Structure-activity relationship homology (SARAH): a conceptual framework for drug discovery in the genomic era. Chem Biol, 6(1), R3-7. Gillet, V. J., Willett, P., & Bradshaw, J. (1998). Identification of biological activity profiles using substructural analysis and genetic algorithms. J Chem Inf Comput Sci, 38(2), 165-179. Glick, M., Robinson, D. D., Grant, G. H., & Richards, W. G. (2002). Identification of ligand binding sites on proteins using a multi-scale approach. J Am Chem Soc, 124(10), 2337-2344. Hadfield, J. A., Ducki, S., Hirst, N., & McGown, A. T. (2003). Tubulin and microtubules as targets for anticancer drugs. Prog Cell Cycle Res, 5, 309-325. Hert, J., Willett, P., Wilton, D. J., Acklin, P., Azzaoui, K., Jacoby, E., et al. (2004). Comparison of topological descriptors for similarity-based virtual screening using multiple bioactive reference structures. Org Biomol Chem, 2(22), 3256-3266. Hochberg, M. C. (2005). COX-2 selective inhibitors in the treatment of arthritis: a rheumatologist perspective. Curr Top Med Chem, 5(5), 443-448. Hofbauer, K. G. (1997, July 5, 1997). Target Identification and Concept Validation in Drug Discovery. Paper presented at the IUPS Satellite Conference 1997: On Designing the Physiome Project, Petrodvoret, St. Petersburg, Russia. Jacoby, E., Schuffenhauer, A., & Floersheim, P. (2003). Chemogenomics knowledgebased strategies in drug discovery. Drug News Perspect, 16(2), 93-102. Khawaja, A. M., & Rogers, D. F. (1996). Tachykinins: receptor to effector. Int J Biochem Cell Biol, 28(7), 721-738. 36 Kitchen, D. B., Stahura, F. L., & Bajorath, J. (2004). Computational techniques for diversity analysis and compound classification. Mini Rev Med Chem, 4(10), 10291039. Klon, A. E., Glick, M., & Davies, J. W. (2004). Application of machine learning to improve the results of high-throughput docking against the HIV-1 protease. J Chem Inf Comput Sci, 44(6), 2216-2224. Klon, A. E., Glick, M., Thoma, M., Acklin, P., & Davies, J. W. (2004). Finding more needles in the haystack: A simple and efficient method for improving highthroughput docking results. J Med Chem, 47(11), 2743-2749. Lagunin, A., Stepanchikova, A., Filimonov, D., & Poroikov, V. (2000). PASS: prediction of activity spectra for biologically active substances. Bioinformatics, 16(8), 747748. Manallack, D. T., Pitt, W. R., Gancia, E., Montana, J. G., Livingstone, D. J., Ford, M. G., et al. (2002). Selecting screening candidates for kinase and G protein-coupled receptor targets using neural networks. J Chem Inf Comput Sci, 42(5), 1256-1262. Melnikova, I., & Golden, J. (2004). Targeting protein kinases. Nat Rev Drug Discov, 3(12), 993-994. Mendelsohn, J., & Baselga, J. (2003). Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J Clin Oncol, 21(14), 27872799. Morgan, H. L. (1965). The generation of a unique machine description for chemical structures - a technique developed at Chemical Abstracts Service. J Chem Doc(5), 107-113. Poroikov, V. V., Filimonov, D. A., Ihlenfeldt, W. D., Gloriozova, T. A., Lagunin, A. A., Borodina, Y. V., et al. (2003). PASS biological activity spectrum predictions in the enhanced open NCI database browser. J Chem Inf Comput Sci, 43(1), 228236. Rabow, A. A., Shoemaker, R. H., Sausville, E. A., & Covell, D. G. (2002). Mining the National Cancer Institute's tumor-screening database: identification of compounds with similar cellular activities. J Med Chem, 45(4), 818-840. Rusinko, A., 3rd, Farmen, M. W., Lambert, C. G., Brown, P. L., & Young, S. S. (1999). Analysis of a large structure/biological activity data set using recursive partitioning. J Chem Inf Comput Sci, 39(6), 1017-1026. Sche, P. P., McKenzie, K. M., White, J. D., & Austin, D. J. (1999). Display cloning: functional identification of natural product receptors using cDNA-phage display. Chem Biol, 6(10), 707-716. Schuffenhauer, A., Zimmermann, J., Stoop, R., van der Vyver, J. J., Lecchini, S., & Jacoby, E. (2002). An ontology for pharmaceutical ligands and its application for in silico screening and library design. J Chem Inf Comput Sci, 42(4), 947-955. Shemetulskis, N. E., Dunbar, J. B., Jr., Dunbar, B. W., Moreland, D. W., & Humblet, C. (1995). Enhancing the diversity of a corporate database using chemical database clustering and analysis. J Comput Aided Mol Des, 9(5), 407-416. Taunton, J., Hassig, C. A., & Schreiber, S. L. (1996). A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272(5260), 408-411. Toledo, L. M., Lydon, N. B., & Elbaum, D. (1999). The structure-based design of ATPsite directed protein kinase inhibitors. Curr Med Chem, 6(9), 775-805. 37 Vigushin, D. M., Ali, S., Pace, P. E., Mirsaidi, N., Ito, K., Adcock, I., et al. (2001). Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo. Clin Cancer Res, 7(4), 971-976. Vigushin, D. M., & Coombes, R. C. (2002). Histone deacetylase inhibitors in cancer treatment. Anticancer Drugs, 13(1), 1-13. Weinstein, J. N., Myers, T. G., O'Connor, P. M., Friend, S. H., Fornace, A. J., Jr., Kohn, K. W., et al. (1997). An information-intensive approach to the molecular pharmacology of cancer. Science, 275(5298), 343-349. Weisstein, E. W. Bayes Theorem [Electronic Version]. MathWorld from http://mathworld.wolfram.com/BayesTheorem.html. Wexler, R. R., Carini, D. J., Duncia, J. V., Johnson, A. L., Wells, G. J., Chiu, A. T., et al. (1992). Rationale for the chemical development of angiotensin II receptor antagonists. Am J Hypertens, 5(12 Pt 2), 209S-220S. Willett, P. (2005). Searching techniques for databases of two- and three-dimensional chemical structures. J Med Chem, 48(13), 4183-4199. Xia, X., Maliski, E. G., Gallant, P., & Rogers, D. (2004). Classification of kinase inhibitors using a Bayesian model. J Med Chem, 47(18), 4463-4470. Xue, L., & Bajorath, J. (2000). Molecular descriptors in chemoinformatics, computational combinatorial chemistry, and virtual screening. Comb Chem High Throughput Screen, 3(5), 363-372. Zhu, L. L., Hou, T. J., Chen, L. R., & Xu, X. J. (2001). 3D QSAR analyses of novel tyrosine kinase inhibitors based on pharmacophore alignment. J Chem Inf Comput Sci, 41(4), 1032-1040. 38 APPENDIX CURRICULUM VITAE NIDHI B. Sc. (chemistry), M.S. (computer applications) Phone: 317-652-1631 Email: khurananidhi@gmail.com SUMMARY Solid grasp on basic sciences through an undergraduate degree in chemistry Extensive software development skills through a master’s degree in computer applications High level of awareness regarding software industry’s practices gained by internships and full time job as software engineer Pursuing a master’s degree in Chemical Informatics at Indiana University, Indianapolis. Worked at Novartis Institutes for BioMedical Research, Cambridge, MA as Cheminformatics Summer Science Intern during summer 2005. SKILLS Cheminformatics Molecular Modeling Languages Databases Platforms Web Technologies Tools & Utilities Others : Pipeline Pilot, Spotfire, ISIS/Base, ChemDraw, BCI Toolkit : Spartan, SYBYL, Macro Model, Maestro : C, JAVA, Visual Basic 6.0 : Oracle 9i, MS-Access : MS Win NT/9x/2K/XP, UNIX, Linux : HTML, XML, VBScript, ASP3.0, JSP, Servlets : Crystal Reports, Matlab : Labware EDUCATIONAL EXPERIENCE MS - Chemical Informatics, Indiana University Purdue University Indianapolis, Indiana. August 2004 – Till date. Expected graduation December 2005 Masters in Computer Application (M.C.A.), Maharishi Dayanand University, Haryana, India. September 2003 Bachelor of Science (B.Sc.)-Chemistry (Honors), Delhi University, New Delhi, India. July 1999 WORK EXPERIENCE Cheminformatics Summer Science Intern: June 2005 – August 2005 - Novartis Institutes for BioMedical Research, Cambridge, MA Worked with Lead Discovery Informatics division (Davies’ group) on Library and Database Annotation Concept for a project to explore the relationship between chemical structures and associated biological activities. Platform: Pipeline Pilot, Spotfire, ISIS/Base Software Engineer - March 2004 – July 2004 - Satyam Computer & Services Ltd., Hyderabad, India Provided offshore client support to Cisco Systems for development of an application that incorporated both Client/Server and n-tier Web Architecture Worked on J2EE platform using Struts framework Documented Use Cases, Test Cases and Design Elements following Satyam’s ISO 9001 quality procedures Successfully completed corporate training modules in OOAD & UML, Java, XML and Quality Assurance Project Intern - January 2003 – June 2003 - Kirloskar Pneumatic Company Ltd., New Delhi, India Involved in development of an Intranet application which automated the Tender Quotation Process of the organization Performed system analysis & design, development and documentation of the system Programming Platform: ASP3.0, VBScript, MS Access 2000 Summer Intern - June 1, 2001 – July 30, 2001. Claas India Ltd., Faridabad, India Involved in the development of a desktop application to manage fixed assets of the corporation. Designed and developed the application single handedly. Programming Platform: VB6.0, Oracle 9i and Crystal Reports 8.0 RELEVANT TRAINING/WORKSHOPS Pipeline Pilot Basic Training Course by SciTegic - June 2005. NCBI workshop on MapViewer – November 2005 AWARDS/FELLOWSHIPS - MDL Elsevier Excellence in Informatics Fellowship for the year 2004 ACDEMIC PROJECTS & PRESENTATIONS Conformational search on sixteen mer of a polymeric material that inhibits HIV-1 reverse transcriptase - Independent study project Fall 2005. Proteomics Laboratory Study and Implementation of workflow on Labware - Class project for Lab Info Management Systems course done in Spring 2005. Tracking system for unsigned Physician Orders - Platform JSP/Oracle 8i - Class project for Informatics Management course done in Spring 2005. Evaluation of BCI Toolkit - Class project for Chemical Information Technology course done in Fall 2004. Quest for Biomarkers - Class project for Introduction to Bioinformatics course done in Fall 2004. DETAILS OF COURSEWORK TAKEN MS in Chemical Informatics 1) Lab Info Management Systems 2) Introduction to Informatics 3) Chemical Information Technology 4) Introduction to Bioinformatics 5) Molecular Modeling 6) Informatics Management 7) Informatics Research & Design Master’s Program in Computer Application (A three-year graduate program) 1) Mathematical foundation of computer science 2) Probability and statistics 3) Computer based numerical & statistical techniques 4) Computer based optimization techniques 5) Computer programming & problem solving 6) Business data processing 7) Combination & graph theory 8) Data & file structure 9) System software and C programming 10) Database management system 11) System analysis & design 12) Operating systems 13) Interactive computer graphics 14) Distributed computer network application 15) Simulation & modeling 16) Artificial intelligence 17) Software engineering 18) Computer organization & assembly language 19) Microprocessor & application 20) Computer architecture 21) Accounts & financial management 22) Organizational structure & personnel management 23) Information system design & implementation Bachelor of Science (with honors in chemistry) (Not a complete list) 1) Inorganic Chemistry 2) Organic Chemistry 3) Physical Chemistry 4) Environmental Chemistry 5) Mathematics 6) Physics