Отчет о белке MurD E.coli в формате doc.
advertisement

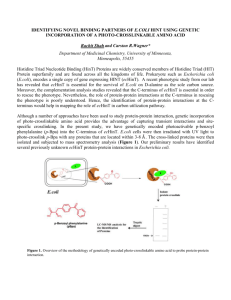
Отчет о белке Murd_ecoli. Полное название белка: Уридиндифосфат-N-ацетилмурамоил-L-аланин: D-глутамат лигаза. Белок относится к классу лигаз и группе белков murCDEF. Локализируется в цитоплазме бактерий Escherichia coli (Таксономия: Bacteria - Proteobacteria Gammaproteobacteria - Enterobacteriales - Enterobacteriaceae - Escherichia). В клетке белок осуществляет один из этапов синтеза клеточной стенки, а именно катализ присоединения глутамата предшественнику нуклеотда УДФ-N- ацетилмурамоилL-аланину по формуле: ATP + UDP-N-acetylmuramoyl-L-alanine + glutamate = ADP + phosphate + UDP-Nacetylmuramoyl-L-alanyl-D- glutamate. Структура белка представляет собой одну цепь, уложенную в структуру, показанную не рисунке (красным на рисунке отмечены альфа-спирали, а желтым бета-тяжи). Длинна этой цепи составляет 438 аминокислотных остатка, а молекулярная масса 46,974 дальтона. С 112 по 118 аминокислотный остаток последовательности лежит центр связывания молекулы АТФ. Данные по количеству альфа-спиралей, бета-тяжей и остальных элементов вторичной структуры в различных базах данных несколько расходятся. В общем случае различают 20 бета-тяжей и 23 альфа-спирали, а также множество различных более коротких элементов вторчной структуры. Полная последовательность белка в формате fasta: >sp|P14900|MURD_ECOLI UDP-N-acetylmuramoylalanine--D-glutamate ligase; MADYQGKNVVIIGLGLTGLSCVDFFLARGVTPRVMDTRMTPPGLDKLPEAVERHTGSL NDEWLMAADLIVASPGIALAHPSLSAAADAGIEIVGDIELFCREAQAPIVAITGSNGKST VTTLVGEMAKAAGVNVGVGGNIGLPALMLLDDECELYVLELSSFQLETTSSLQAVAAT ILNVTEDHMDRYPFGLQQYRAAKLRIYENAKVCVVNADDALTMPIRGADERCVSFGV NMGDYHLNHQQGETWLRVKGEKVLNVKEMKLSGQHNYTNALAALALADAAGLPRA SSLKALTTFTGLPHRFEVVLEHNGVRWINDSKATNVGSTEAALNGLHVDGTLHLLLGG DGKSADFSPLARYLNGDNVRLYCFGRDGAQLAALRPEVAEQTETMEQAMRLLAPRVQ PGDMVLLSPACASLDQFKNFEQRGNEFARLAKELG *Большинство данных о белке и некоторые рисунки взяты с сайта UniProt (http://www.uniprot.org), PubSUM EMBL (http://www.ebi.ac.uk/pdbsum) и RSCB PDB (http://www.rcsb.org) Подробнее о белке можно узнать из записей с сайтов UniProt: http://www.uniprot.org/uniprot/P14900 А также RSCB PDB http://www.rcsb.org/pdb/explore.do?structureId=1EEH Статьи о белке (англ). Статьи представлены в виде названий, или аннотаций. 1. Nucleotide sequence involving murD and an open reading frame ORF-Y spacing murF and ftsW in Escherichia coli. Ikeda M., Wachi M., Ishino F., Matsuhashi M. Nucleic Acids Res. 18:1058-1058(1990) 2. Nucleotide sequence of the murD gene encoding the UDP-MurNAc-L-Ala-D-Glu synthetase of Escherichia coli. Mengin-Lecreulx D., van Heijenoort J. Nucleic Acids Res. 18:183-183(1990) 3. Systematic sequencing of the Escherichia coli genome: analysis of the 0-2.4 min region. Yura T., Mori H., Nagai H., Nagata T., Ishihama A., Fujita N., Isono K., Mizobuchi K., Nakata A. A contiguous 111,402-nucleotide sequence corresponding to the 0 to 2.4 min region of the E. coli chromosome was determined as a first step to complete structural analysis of the genome. The resulting sequence was used to predict open reading frames and to search for sequence similarity against the PIR protein database. A number of novel genes were found whose predicted protein sequences showed significant homology with known proteins from various organisms, including several clusters of genes similar to those involved in fatty acid metabolism in bacteria (e.g., betT, baiF) and higher organisms, iron transport (sfuA, B, C) in Serratia marcescens, and symbiotic nitrogen fixation or electron transport (fixA, B, C, X) in Azorhizobium caulinodans. In addition, several genes and IS elements that had been mapped but not sequenced (e.g., leuA, B, C, D) were identified. We estimate that about 90 genes are represented in this region of the chromosome with little spacer. Nucleic Acids Res. 20:3305-3308(1992) 4. The complete genome sequence of Escherichia coli K-12. Blattner F.R., Plunkett G. III, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Gregor J., Davis N.W., Kirkpatrick H.A., Goeden M.A., Rose D.J., Mau B., Shao Y. The 4,639,221-base pair sequence of Escherichia coli K-12 is presented. Of 4288 protein-coding genes annotated, 38 percent have no attributed function. Comparison with five other sequenced microbes reveals ubiquitous as well as narrowly distributed gene families; many families of similar genes within E. coli are also evident. The largest family of paralogous proteins contains 80 ABC transporters. The genome as a whole is strikingly organized with respect to the local direction of replication; guanines, oligonucleotides possibly related to replication and recombination, and most genes are so oriented. The genome also contains insertion sequence (IS) elements, phage remnants, and many other patches of unusual composition indicating genome plasticity through horizontal transfer. Science 277:1453-1474(1997) 5. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Hayashi K., Morooka N., Yamamoto Y., Fujita K., Isono K., Choi S., Ohtsubo E., Baba T., Wanner B.L., Mori H., Horiuchi T. With the goal of solving the whole-cell problem with Escherichia coli K-12 as a model cell, highly accurate genomes were determined for two closely related K-12 strains, MG1655 and W3110. Completion of the W3110 genome and comparison with the MG1655 genome revealed differences at 267 sites, including 251 sites with short, mostly single-nucleotide, insertions or deletions (indels) or base substitutions (totaling 358 nucleotides), in addition to 13 sites with an insertion sequence element or defective prophage in only one strain and two sites for the W3110 inversion. Direct DNA sequencing of PCR products for the 251 regions with short indel and base disparities revealed that only eight sites are true differences. The other 243 discrepancies were due to errors in the original MG1655 sequence, including 79 frameshifts, one amino-acid residue deletion, five amino-acid residue insertions, 73 missense, and 17 silent changes within coding regions. Errors in the original MG1655 sequence (<1 per 13,000 bases) were mostly within portions sequenced with out-dated technology based on radioactive chemistry. Mol. Syst. Biol. 2:E1-E5(2006) 6. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. Ikeda M., Sato T., Wachi M., Jung H.K., Ishino F., Kobayashi Y., Matsuhashi M. The Escherichia coli cell division gene ftsW (2 min) was cloned and sequenced. It encodes a hydrophobic protein(s) with 414 and/or 384 amino acid residues. The deduced amino acid sequence and the hydropathy profile of the protein showed high homology with those of the E. coli RodA protein functioning in determination of the cell shape and the Bacillus subtilis SpoVE protein functioning in spore formation. Probably similar functional membrane proteins are involved in these three cell cycle process. J. Bacteriol. 171:6375-6378(1989) 7. Over-production, purification and properties of the uridine diphosphate Nacetylmuramoyl-L-alanine:D-glutamate ligase from Escherichia coli. Pratviel-Sosa F., Mengin-Lecreulx D., van Heijenoort J. The UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase of Escherichia coli was overproduced in strains that harbour recombinant plasmids bearing the murD gene under the control of the lac or PR promoter. Purification to homogeneity was achieved by a two-step procedure from a 181-fold over-producing strain. The N-terminal sequence of the purified protein was determined and correlated with the nucleotide sequence of the murD gene. The purified activity was highly dependent on the concentration of potassium phosphate and Mg2+. The enzyme also catalysed the reverse reaction. The Km values for UDP-N-acetylmuramoyl-L-alanine; Dglutamate and ATP/Mg2+ were estimated at 7.5, 55 and 138 microM, respectively. Under the most optimal in vitro conditions determined, a turnover number of 931 min-1 was estimated. When considering the plasmid-free parental strain, the copy number of the murD gene product was not more than 1000.cell-1. 8. Crystal structure of UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase from Escherichia coli. Bertrand J.A., Auger G., Fanchon E., Martin L., Blanot D., van Heijenoort J., Dideberg O. UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase (MurD) is a cytoplasmic enzyme involved in the biosynthesis of peptidoglycan which catalyzes the addition of D-glutamate to the nucleotide precursor UDP-N-acetylmuramoyl-L-alanine (UMA). The crystal structure of MurD in the presence of its substrate UMA has been solved to 1.9 A resolution. Phase information was obtained from multiple anomalous dispersion using the K-shell edge of selenium in combination with multiple isomorphous replacement. The structure comprises three domains of topology each reminiscent of nucleotide-binding folds: the N- and C-terminal domains are consistent with the dinucleotide-binding fold called the Rossmann fold, and the central domain with the mononucleotide-binding fold also observed in the GTPase family. The structure reveals the binding site of the substrate UMA, and comparison with known NTP complexes allows the identification of residues interacting with ATP. The study describes the first structure of the UDP-N-acetylmuramoyl-peptide ligase family. EMBO J. 16:3416-3425(1997) 9. 'Open' structures of MurD: domain movements and structural similarities with folylpolyglutamate synthetase. Bertrand J.A., Fanchon E., Martin L., Chantalat L., Auger G., Blanot D., van Heijenoort J., Dideberg O. UDP-N-acetylmuramoyl-l-alanine:d-glutamate (MurD) ligase catalyses the addition of dglutamate to the nucleotide precursor UDP-N-acetylmuramoyl-l-alanine (UMA). The crystal structures of Escherichia coli in the substrate-free form and MurD complexed with UMA have been determined at 2.4 A and 1.88 A resolution, respectively. The MurD structure comprises three domains each of a topology reminiscent of nucleotide-binding folds. In the two structures the C-terminal domain undergoes a large rigid-body rotation away from the N-terminal and central domains. These two "open" structures were compared with the four published "closed" structures of MurD. In addition the comparison reveals which regions are affected by the binding of UMA, ATP and d-Glu. Also we compare and discuss two structurally characterized enzymes which belong to the same ligase superfamily: MurD and folylpolyglutamate synthetase (FGS). The analysis allows the identification of key residues involved in the reaction mechanism of FGS. The determination of the two "open" conformation structures represents a new step towards the complete elucidation of the enzymatic mechanism of the MurD ligase. J. Mol. Biol. 301:1257-1266(2000) Мартынов Александр, 2009 г.