Paired and Matched Binary Data
advertisement

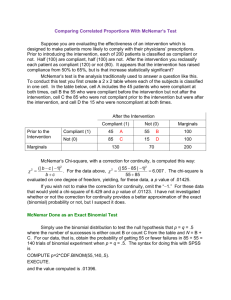
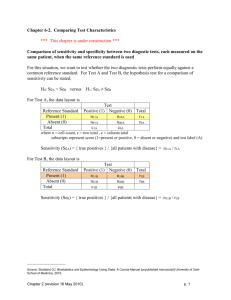
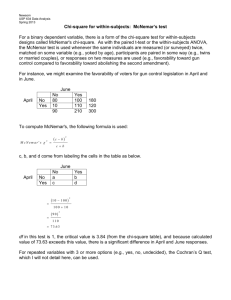
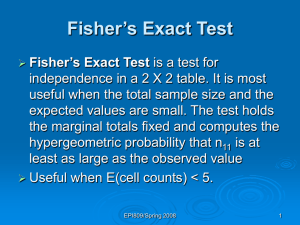
Note 17 Paired and Matched Binary Data The data here are from a clinical trial of an allergy medication. Participants recorded use of decongestants before starting on study medication and then after using study medication for four weeks. deco0 records usage (0/1) before medication, deco1 after medication. [17.1] . use hepp-patient, clear . list +--------------------------+ | id deco0 deco1 n | |--------------------------| 1. | 1 0 0 237 | 2. | 2 0 1 57 | 3. | 3 1 0 71 | 4. | 4 1 1 14 | +--------------------------+ The first command below shows an incorrect analysis, since the data are actually paired. (That is, the test for independence here takes no note of the fact that there are actually 379 pairs of measurements represented here.) The correct analysis uses McNemar’s test, which looks only at the diagonals of the 2 × 2 table, the only cells which are informative about how the before and after probabilities differ. [17.2] . ta deco0 deco1 [fw=n], chi2 exact ** WRONG ** Decongesta | nt before | Decongestant after rx rx | 0 1 | Total -----------+----------------------+---------0 | 237 57 | 294 1 | 71 14 | 85 -----------+----------------------+---------Total | 308 71 | 379 Pearson chi2(1) = Fisher’s exact = 1-sided Fisher’s exact = 0.3686 Pr = 0.544 ** WRONG ** 0.637 ** WRONG ** 0.332 ** WRONG ** The following commands do a correct analysis. The first does McNemar’s test using the exact binomial probabilities, while the next two use the normal approximation. When the z value computed below is squared, it is very close to McNemar’s χ2 test, which is calculated in the last display. 1 NOTE 17. PAIRED AND MATCHED BINARY DATA 2 [17.3] . bitesti 128 71 .5 N Observed k Expected k Assumed p Observed p -----------------------------------------------------------128 71 64 0.50000 0.55469 Pr(k >= 71) = 0.125220 Pr(k <= 71) = 0.907659 Pr(k <= 57 or k >= 71) = 0.250440 (one-sided test) (one-sided test) (two-sided test) [17.4] . display "z = " (71/128 - 0.5)/sqrt(71*57/(128*128*128)) z = 1.2449056 [17.5] . display (1.2449056)^2 " 1.54979 .21316645 " 2*(1-normprob(1.2449056)) [17.6] . display "McNemar’s chi-squared statistic = " ((71-57)^2)/(71+57) McNemar’s chi-squared statistic = 1.53125 The mcc command analyzes a matched case-control study, and is the way to calculate McNemar’s test for a paired binomial data set in Stata. Unfortunately, one must reinterpret the labels that Stata uses, as this is obviously not a case-control study. You can think of treatment status as defining case vs. control, so that cases correspond to the “after” measurement and controls to the “before.” [17.7] . mcc deco1 deco0 [fw=n] | Controls | Cases | Exposed Unexposed | Total -----------------+------------------------+-----------Exposed | 14 57 | 71 Unexposed | 71 237 | 308 -----------------+------------------------+-----------Total | 85 294 | 379 McNemar’s chi2(1) = 1.53 Prob > chi2 = 0.2159 Exact McNemar significance probability = 0.2504 Proportion with factor Cases .1873351 Controls .2242744 --------difference -.0369393 ratio .8352941 rel. diff. -.047619 odds ratio .8028169 [95% Conf. Interval] --------------------.0979673 .0240887 .6278769 1.111231 -.1248173 .0295792 .5564015 1.153877 (exact) NOTE 17. PAIRED AND MATCHED BINARY DATA 3 More generally, we could have several “controls” for each “case”, in which case the approach above cannot readily be used. The clogit command does conditional logistic regression, which is a generalization of McNemar’s test. The data need to be in long format for this command, so we start by having Stata reshape the data set. [17.8] . reshape long deco, i(id) j(after) (note: j = 0 1) Data wide -> long ----------------------------------------------------------------------------Number of obs. 4 -> 8 Number of variables 4 -> 4 j variable (2 values) -> after xij variables: deco0 deco1 -> deco ----------------------------------------------------------------------------[17.9] . list, sep(0) 1. 2. 3. 4. 5. 6. 7. 8. [17.10] +-------------------------+ | id after deco n | |-------------------------| | 1 0 0 237 | | 1 1 0 237 | | 2 0 0 57 | | 2 1 1 57 | | 3 0 1 71 | | 3 1 0 71 | | 4 0 1 14 | | 4 1 1 14 | +-------------------------+ . clogit after deco [fw=n], group(id) nolog or Conditional (fixed-effects) logistic regression Log likelihood = -261.93562 Number of obs LR chi2(1) Prob > chi2 Pseudo R2 = = = = 758 1.53 0.2155 0.0029 -----------------------------------------------------------------------------after | Odds Ratio Std. Err. z P>|z| [95% Conf. Interval] -------------+---------------------------------------------------------------deco | .8028169 .1427759 -1.23 0.217 .5665467 1.13762 ------------------------------------------------------------------------------ NOTE 17. PAIRED AND MATCHED BINARY DATA 4 There is yet one more way to carry out these calculations. McNemar’s test can be thought of as a test of symmetry in the 2 × 2 table about the main diagonal. Sometimes we have paired observations of ordered categories. In the example below, antihistamine use before and after treatment is recorded in patients receiving an allergy treatment. [17.11] . use hepp-anti-full, clear [17.12] . symmetry before after [fw=n] ----------------------------------------------------------------------| after before | None Occasional Daily low Daily full Total -----------+----------------------------------------------------------None | 199 56 8 5 268 Occasional | 34 16 2 0 52 Daily low | 15 6 3 1 25 Daily full | 25 7 2 4 38 | Total | 273 85 15 10 383 ----------------------------------------------------------------------chi2 df Prob>chi2 -----------------------------------------------------------------------Symmetry (asymptotic) | 30.17 6 0.0000 Marginal homogeneity (Stuart-Maxwell) | 30.11 3 0.0000 -----------------------------------------------------------------------In this case, considerable information is lost if the data are simply reduced to antihistamine use (any or none). In the 2 × 2 case, both symmetry tests are equal to the McNemar’s χ2 test. [17.13] . symmetry antibefore antiafter [fw=n], exact ------------------------------antibefor | antiafter e | 0 1 Total ----------+-------------------0 | 199 69 268 1 | 74 41 115 | Total | 273 110 383 ------------------------------chi2 df Prob>chi2 -----------------------------------------------------------------------Symmetry (asymptotic) | 0.17 1 0.6759 Marginal homogeneity (Stuart-Maxwell) | 0.17 1 0.6759 -----------------------------------------------------------------------Symmetry (exact significance probability) 0.7381