Comparing Test Characteristics
advertisement
Chapter 6-2. Comparing Test Characteristics
*** This chapter is under construction ***
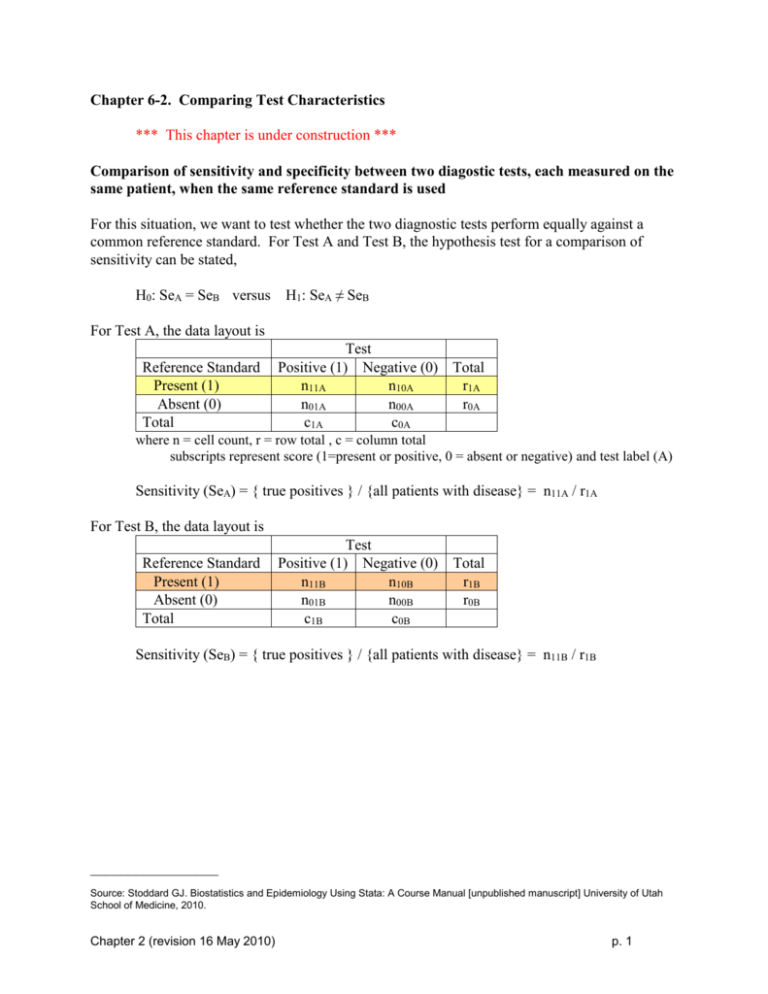
Comparison of sensitivity and specificity between two diagostic tests, each measured on the
same patient, when the same reference standard is used
For this situation, we want to test whether the two diagnostic tests perform equally against a
common reference standard. For Test A and Test B, the hypothesis test for a comparison of
sensitivity can be stated,
H0: SeA = SeB versus
H1: SeA ≠ SeB
For Test A, the data layout is
Reference Standard
Present (1)
Absent (0)
Total
Test
Positive (1) Negative (0) Total
n11A
n10A
r1A
n01A
n00A
r0A
c1A
c0A
where n = cell count, r = row total , c = column total
subscripts represent score (1=present or positive, 0 = absent or negative) and test label (A)
Sensitivity (SeA) = { true positives } / {all patients with disease} = n11A / r1A
For Test B, the data layout is
Reference Standard
Present (1)
Absent (0)
Total
Test
Positive (1) Negative (0) Total
n11B
n10B
r1B
n01B
n00B
r0B
c1B
c0B
Sensitivity (SeB) = { true positives } / {all patients with disease} = n11B / r1B
_________________
Source: Stoddard GJ. Biostatistics and Epidemiology Using Stata: A Course Manual [unpublished manuscript] University of Utah
School of Medicine, 2010.
Chapter 2 (revision 16 May 2010)
p. 1
We see that all information for sensitivity for each test is contained in the first row, where the
first row of each table is the true presence of disease as identified by the common reference
standard. For a paired comparison of sensitivity, then, all we need are the cell counts in these
rows, combined into a paired crosstabulation table.
The paired data layout for a sensitivity comparison is
Test A
Test B
Positive (1) Negative (0)
Positive (1)
m11
m10
Negative (0)
m01
m00
Total
n11A
n10A
Total
n11B
n10B
Where the cell counts, the m’s, simply fill in by the crosstabulation procedure.
Since the data are not independent, being repeated measures on the same patient (both tests done
on same patient), we must apply a paired proportions comparision. To compare sensitivity, we
simply apply the McNemar test, which is the standard way to compare two paired binary
variables expressed in this paired data layout (Lachenbruch and Lynch, 1998; Zhou et al, 2002,
pp.166-169).
paired data layout for a sensitivity comparison
Test A
Test B
Positive (1) Negative (0) Total
Positive (1)
m11
m10
n11B
Negative (0)
m01
m00
n10B
Total
n11A
n10A
The McNemar test is commonly referred to as the “McNemar change test”, as it only uses
information from the discordant pairs (the cells where the two diagnostic tests are different).
It is simply a chi-square test (Siegel and Castellan, 1988, p.76) expressed as,
2
df 1
(m10 m01 ) 2
m10 m01
Small Expected Frequencies
The chi-square test requires a sufficiently large sample size to provide an accurate p value. The
rule-of-thumb for the McNemar test version of the chi-square test is that when (m10 + m01) < 10,
the exact form of the test should be used (Siegel and Castellan, 1988, p.79). Since the data are
paired, the Fisher’s exact test is not appropriate, and so the binomial test is used. In Stata, this
binomial test is labeled “Exact McNemar”.
Chapter 2 (revision 16 May 2010)
p. 2
Specificity
The comparison of specificity is done is an anologous fashion. The hypothesis becomes
H0: SpA = SpB versus
H1: SpeA ≠ SpB
For Test A, the data layout is
Test
Gold Standard Positive (1) Negative (0)
Present (1)
n11A
n10A
Absent (0)
n01A
n00A
Total
c1A
c0A
Total
r1A
r0A
where n = cell count, r = row total , c = column total
subscripts represent score (1=present or positive, 0 = absent or negative) and test label (A)
Specificity (SpA) = { true negative } / {all patients without disease} = n00A / r0A
For Test B, the data layout is
Gold Standard Positive (1)
Present (1)
n11B
Absent (0)
n01B
Total
c1B
Test
Negative (0)
n10B
n00B
c0B
Total
r1B
r0B
Specificity (SpB) = { true negative } / {all patients without disease} = n00B / r0B
We see that all information for specificity for each test is contained in the second row, where the
second row of each table is the true absence of disease as identified by the common reference
standard. For a paired comparison of specificity, then, all we need are the cell counts in these
rows, combined into a paired crosstabulation table.
The paired data layout for a specificity comparison is
Test A
Test B
Positive (1) Negative (0) Total
Positive (1)
m11
m10
n01B
Negative (0)
m01
m00
n00B
Total
n01A
n00A
Where the cell counts, the m’s, simply fill in by the crosstabulation procedure.
Then, McNemar’s test is applied in an identical way to the sensitivity comparison.
Chapter 2 (revision 16 May 2010)
p. 3
Protocol Suggestion
For comparison of sensitivity and specificity between two diagnostic tests, you could describe the
statistical method as:
Within the same patients, both Test A and Test B will be compared to a common Test C
gold standard and test characteristics will be calculated. The sensitivity between Test A
and Test B will be compared using a McNemar test, or exact McNemar test, as
appropriate [Lachenbruch and Lynch, 1998]. The specificity will similarly be compared.
Example
We will use the CASS dataset (see Appendix 1 for references). These data come from the
coronary artery surgery study (CASS). In a cohort study of N=1465 men undergoing coronary
arteriography (the gold standard) for suspected or probable coronary heart disease, both an
exercise stress test (EST) and chest pain history (CPH) were recorded. The data are coded as
cad
est
cph
coronary artery disease (gold standard), 1 = yes, 0 = no
exercise stress test (diagnostic test for CAD), 1 = positive, 0 = negative
chest pain history (diagnostic test for CAD), 1 = positive, 0 = negative
Reading in the data into Stata,
File
Open
Find the directory where you copied the course CD:
Find the subdirectory datasets & do-files
Single click on cass.dta
Open
use "C:\Documents and Settings\u0032770.SRVR\Desktop\
Biostats & Epi With Stata\datasets & do-files\cass.dta", clear
*
which must be all on one line, or use:
cd "C:\Documents and Settings\u0032770.SRVR\Desktop\"
cd "Biostats & Epi With Stata\datasets & do-files"
use cass.dta, clear
To obtain the sensitivity and specificity for est, we use the diagt command, which is not available
from the Stata menu bar.
Chapter 2 (revision 16 May 2010)
p. 4
If you have not already updated your Stata to include it, then while connected to the internet, use
findit diagt
Search of official help files, FAQs, Examples, SJs, and STBs
SJ-4-4
sbe36_2 . . . . . . . . . . . . . . . . . . Software update for diagt
(help diagt if installed) . . . . . . . . . . P. T. Seed and A. Tobias
Q4/04
SJ 4(4):490
new options added to diagt
Click on the sbe36_2 link, or a later version if one appears, to install the diagt command.
To obtain the test characteristics to est, use
diagt cad est
Coronary |
artery | Exercise Stress test
disease |
Pos.
Neg. |
Total
-----------+----------------------+---------Abnormal |
815
208 |
1,023
Normal |
115
327 |
442
-----------+----------------------+---------Total |
930
535 |
1,465
[95% Confidence Interval]
--------------------------------------------------------------------------Prevalence
Pr(A)
70%
67%
72.2%
--------------------------------------------------------------------------Sensitivity
Pr(+|A)
79.7%
77.1%
82.1%
Specificity
Pr(-|N)
74%
69.6%
78%
---------------------------------------------------------------------------
To obtain the sensitivity for cph,
diagt cad cph
Coronary |
artery | Chest pain history
disease |
Pos.
Neg. |
Total
-----------+----------------------+---------Abnormal |
969
54 |
1,023
Normal |
245
197 |
442
-----------+----------------------+---------Total |
1,214
251 |
1,465
[95% Confidence Interval]
--------------------------------------------------------------------------Prevalence
Pr(A)
70%
67%
72.2%
--------------------------------------------------------------------------Sensitivity
Pr(+|A)
94.7%
93.2%
96%
Specificity
Pr(-|N)
44.6%
39.9%
49.3%
---------------------------------------------------------------------------
Chapter 2 (revision 16 May 2010)
p. 5
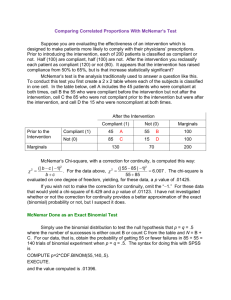
To compute the McNemar test for the sensitivity comparison between the two diagnostic tests,
we restrict the data to the disease present rows, using an if qualifier
mcc cph est if cad==1
| Controls
|
Cases
|
Exposed
Unexposed |
Total
-----------------+------------------------+-----------Exposed |
786
183 |
969
Unexposed |
29
25 |
54
-----------------+------------------------+-----------Total |
815
208 |
1023
McNemar's chi2(1) =
111.87
Prob > chi2 = 0.0000
Exact McNemar significance probability
= 0.0000
We see that the sum of the discordent pairs, 183+29 > 10, so that the sample size is large enough
to provide an accurate chi-square test p value. Therefore, we report the chi-square version of
McNemar’s test (p < 0.001). If, however, the discordant pairs had summed to a number < 10, we
would report the Exact McNemar test (p < .001).
Unfortunately, the variables are labeled cases and controls, which is rather confusing. It is
labelled this way because the McNemar test is part of the epitab suite of commands (the
epidemiology statistical procedures). To verify which variable represents cases, and which
represents controls, we can use,
tab cph est if cad==1
Chest pain | Exercise Stress test
history |
0. neg
1. pos |
Total
-----------+----------------------+---------0. neg |
25
29 |
54
1. pos |
183
786 |
969
-----------+----------------------+---------Total |
208
815 |
1,023
This output has the row and column variables consistent with the mcc command, but displays it
in ascending sort order.
Chapter 2 (revision 16 May 2010)
p. 6
To compute the McNemar test for the specificity comparison between the two diagnostic tests,
we restrict the data to the disease absent rows, using an if qualifier
mcc cph est if cad==0
tab cph est if cad==0
| Controls
|
Cases
|
Exposed
Unexposed |
Total
-----------------+------------------------+-----------Exposed |
69
176 |
245
Unexposed |
46
151 |
197
-----------------+------------------------+-----------Total |
115
327 |
442
McNemar's chi2(1) =
76.13
Prob > chi2 = 0.0000
Exact McNemar significance probability
= 0.0000
Chest pain | Exercise Stress test
history |
0. neg
1. pos |
Total
-----------+----------------------+---------0. neg |
151
46 |
197
1. pos |
176
69 |
245
-----------+----------------------+---------Total |
327
115 |
442
Comparing ROCs
Protocol Suggestion For Comparison of ROCs Using roccomp Is Used
In Stata, the method for comparing two ROCs, as programmed in the roccomp command, is
described by DeLong et al (1988). You could describe this in your protocol as,
The area under the receiver operating characteristic (ROC) curves were computed. For
comparisons of the ROC from different prediction rules, or prognostic models, using a
common reference standard, the method of DeLong et al (1988) was used.
---DeLong ER, Delong DM, Clark-Pearson DL. Comparing the areas under two or more
correlated receiver operating characteristic curves: a nonparametric approach. Biometrics
1988;44(3):837-845.
Chapter 2 (revision 16 May 2010)
p. 7
References
DeLong ER, Delong DM, Clark-Pearson DL. (1988). Comparing the areas under two or more
correlated receiver operating characteristic curves: a nonparametric approach. Biometrics
44(3):837-845.
Lachenbruch PA, Lynch C. (1998). Assessing screening tests: extensions of McNemar’s test.
Statist Med 17:2207-2217.
Siegel S, Castellan NH Jr. (1988). Nonparametric Statistics for the Behavioral Sciences. 2nd ed.
New York, McGraw Hill.
Zhou X-H, Obuchowski NA, McClish DK. (2002). Statistical Methods in Diagnostic Medicine.
New York, John Wiley & Sons.
Chapter 2 (revision 16 May 2010)
p. 8