Live-cell visualization of transmembrane protein oligomerization and
advertisement

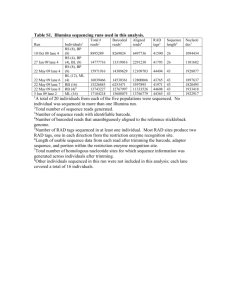
Biosci. Rep. (2012) / 32 / 333–343 (Printed in Great Britain) / doi 10.1042/BSR20110100 SUPPLEMENTARY ONLINE DATA Live-cell visualization of transmembrane protein oligomerization and membrane fusion using two-fragment haptoEGFP methodology Derek J. QUINN*, Neil V. MCFERRAN*, John NELSON* and W. Paul DUPREX†1 *School of Biological Sciences, Queen’s University of Belfast, 97 Lisburn Road, Belfast BT9 7BL, U.K., and †Department of Microbiology, Boston University School of Medicine, Boston, MA 02118, U.S.A. Figure S1 Quantification of the fluorescence signal from Vero cells transfected with pN157 (6)zip and pzip(4)C158 haptoEGFP split point constructs fused to known interacting leucine zippers (A) Generated fluorescence was read in a Tecan fluorescent plate reader (excitation, 485 nm and emission, 535 nm). The results are represented as equivalent sgGFP expressed in femtomoles; also shown are negative controls demonstrating that the generated fluorescent signal from reconstituted haptoEGFP fragments is driven only by the specific interaction of the leucine zippers. (B) Corresponding bar chart showing the quantified fluorescent signal for Vero cells transfected with pN172 (6)zip and pzip(4)C173 haptoEGFP split point constructs. 1 To whom correspondence should be addressed (email pduprex@bu.edu) www.bioscirep.org / Volume 32 (3) / Pages 333–343 D. J. Quinn and others Figure S2 Immunoblotting of Vero cells transfected with various complementary haptoEGFP constructs spilt at several different loop regions and fused to a known interacting pair of leucine zippers Detection of expressed proteins was carried out by immunoblotting with a mixture of two anti-EGFP rabbit polyclonal antisera. (A) HaptoEGFP construct pN142 (6)zip- and pzip(4)C143 + -transfected cells. Lane 1, SeeBlue plus 2 molecular-mass markers; lane 2, mock-transfected Vero cells; lane 3, hapto fusion construct pN142 (6)zip-transfected cells; lane 4, hapto fusion construct pzip(4)C143 -transfected cells); lane 5, hapto fusion constructs pN142 (6)zip and pzip(4)C143 -transfected cells; lane 6, pNPSA(EGFP)-transfected cells; lane 7, recombinant MVeGFP virus-infected cells; lane 8, SeeBlue plus 2 molecular-mass markers. (B) Hapto construct pN157 (6)zip- and pzip(4)C158 -transfected cells. Lane 1, SeeBlue plus 2 molecular-mass markers; lane 2, mock-transfected Vero cells; lane 3, hapto fusion construct pN157 (6)zip-transfected cells; lane 4, hapto fusion construct pzip(4)C158 -transfected cells; lane 5, hapto fusion constructs pN157 (6)zip and pzip(4)C158 -transfected cells; lane 6, pNPSA(EGFP)-transfected cells; lane 7, recombinant MVeGFP virus-infected cells; lane 8, SeeBlue plus 2 molecular-mass markers. (C) Hapto construct pN172 (6)zip- and pzip(4)C173 -transfected cells. Lane 1, SeeBlue plus 2 molecular-mass markers; lane 2, mock-transfected Vero cells; lane 3, hapto fusion construct pN172 (6)zip-transfected cells; lane 4, hapto fusion construct pzip(4)C173 -transfected cells; lane 5, hapto fusion constructs pN172 (6)zip- and pzip(4)C173 -transfected cells; lane 6, pNPSA(EGFP)-transfected cells; lane 7, recombinant MVeGFP virus-infected cells; lane 8, SeeBlue plus 2 molecular-mass markers. (D) Hapto construct pN190 (6)zip- and pzip(4)C191 -transfected cells. Lane 1, SeeBlue plus 2 molecular-mass markers; lane 2, mock-transfected Vero cells; lane 3, hapto fusion construct pN190 (6)zip-transfected cells; lane 4, hapto fusion construct pzip(4)C191 -transfected cells; lane 5, hapto fusion constructs pN190 (6)zip- and pzip(4)C191 -transfected cells; lane 6, pNPSA(EGFP)-transfected cells; lane 7, recombinant MVeGFP virus-infected cells; lane 8, SeeBlue plus 2 molecular-mass markers. Figure S3 Quantification of the fluorescence signal from Vero cells transfected with various MV-H and MV-F haptoEGFP fusion constructs Generated fluorescence signal was read on a Tecan fluorescent plate reader (excitation 485 nm, and emission 535 nm). The results are represented as equivalent sgGFP expressed in femtomoles. Also shown are negative controls where complementary MV-H or MV-F haptoEGFR fusion constructs have been replaced with those fused to a leucine zipper, demonstrating that the fluorescent signal is generated only by the specific interaction of the MV glycoproteins in either homo-oligomeric or hetero-oligomeric complexes. The final bar shows the positive control level of fluorescence obtained with two complementary leucine zipper fusions. The increased level of fluorescence observed with the combined MV-H/MV-F haptoEGFP constructs, as compared with the experiments involving a single viral protein fusion, reflects the potential for viral-protein-driven non-fluorogenic homotrimerization of non-complementary haptoEGFPs, such as trimeric [N157 (16)H]3 . In the case of the N157 (16)H/F(14)C158 experiment, all H–F glycoprotein associations will lead to fluorescence. Received 30 August 2011/22 November 2011; accepted 2 March 2012 Published as Immediate Publication 2 March 2012, doi 10.1042/BSR20110100 .......................................................................................................................................................................................................................................................................................................................................................................... C The Authors Journal compilation C 2012 Biochemical Society