Phylogenetics
advertisement

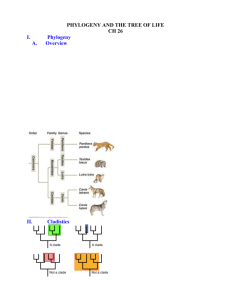
Phylogenetics We’re now moving into the realm of macroevolution: relationships among divergent species… Monophyletic Paraphyletic group Phylogeny (=tree): graphical representation of the historical relationships among species or higher taxonomic groups A B D A C B speciation C speciation A and B are sister group to C and D: diverged from shared common ancestor D speciation These phylogenies show the same relationship. Reptiles are paraphyletic Vertebrate phylogeny: Monophyletic: group derived from one common ancestor and that represents all descendents of that common ancestor (= clade). Paraphyletic: group derived from one common ancestor and that represents some but not all descendents of that common ancestor. Polyphyletic: group does not share a single common ancestor in the time frame under consideration. Basic idea of phylogenetic inference: traits (=characters) that are more similar reflect more recent common ancestry Homology: trait similarity reflecting shared common ancestry ray finned fish lungfish e.g., salamanders Same underlying skeletal elements Mammalian forelimb Synapomorphy: shared, derived form of a trait frogs lizards (e.g., human/cat: forelimb bone ratio) snakes turtles Reptiles crocodiles birds mammals human cat whale bat Autapomorphy: derived form of a trait unique to a group (e.g., bat phalanges) 1 Nested synapomorphies define a phylogeny For a homologous gene, mutations define synapomorphies: e.g., portion of Cytochrome c gene sequence… Synapomorphy: shared, derived form of a trait Autapomorphy: derived form of a trait unique to a group (not phylogenetically informative) Other birds Species 1 Species 2 Species 3 Perching birds (passerines) Finches T at position 7: synapomorphy for species 2/3 12 tail feathers, thick beak: finch synapomorphy 3 front toes, 1 back toe: passerine synapomorphy Phylogeny defined solely by synapomorphies: cladogram Synapomorphy: shared, derived trait… We’ve already seen an example of outgroup rooting: Q: For a variable trait, how do you know which form is derived? Outgroup comparison demonstrates that female preference predates long tails. A: Outgroup comparison: compare those species of interest (=ingroup) to closely related species (=outgroup) Female preference for long tails (P+) is a synapomorphy that groups species 1 and 2. The outgroup roots the tree: indicates polarity (direction) of change: ingroup a* a* a Intersexual selection: sensory bias explanation for female choice outgroup a Species 1 a Species 2 a a* Polytomy (unresolved branch) a Outgroup tells us that a* is derived, so a* species are a monophyletic group. P+: females prefer long tails T+: males possess long tails 2 Homology vs. Homoplasy Homoplasy: trait similarity that does not reflect common ancestry Homoplasy: trait similarity that does not reflect common ancestry: Can arise by evolutionary convergence or evolutionary reversal 1) Evolutionary reversal back to the ancestral form: Fish eye Arises by: Octopus eye Homoplasy: trait similarity that does not reflect common ancestry: DNA sequences: only 4 variants, so homoplasy easily obscures divergence. Especially for mutational hotspots: repeated evolution at the same sites Arises by: 2) Evolutionary convergence: independent evolution of a derived trait % divergence ATTGCTATTC ATTGCTTTTC ATTGCTCTTC ATTGCTTTTC T to C ATTGCTCTTC T to C # nucleotide changes To sp. 1 sp. 2 ATCGATGCAC TTCGATGCAG 20% 2 T1 sp. 1 sp. 2 ATCGATGCAC GTCGATGCAC 10% 2+2=4 At time To: 2 substitutions = 20% divergence At time T1: there have been 2 more substitutions, but now there is only 10% divergence 3 DNA sequences: multiple hits (mutations) over enough time will ultimately obscure any phylogenetic signal. % sequence divergence II e.g., Dog-like mammalian carnivores Marsupial: Varies by gene, depending on mutation rate I Analagous features arise through evolutionary convergence Placental: III 75% Time since divergence from common ancestor Multiple mutations will ultimately randomize sequences: for 4 nucleotides, expect 25% similarity by chance alone Tasmanian wolf (Thylacinus cynocephalus) Dingo (Canis lupis) Placental mammals I. Divergence obeys molecular clock: phylogenetic inference straightforward II. Need statistical correction for multiple hits III. Saturation: no phylogenetic information Evolutionary convergence in beak shape: distantly related nectar feeding birds Marsupials Evolutionary convergence in plant form: cool tropics Colombia South Africa Hawaii Tasmania 4 Evolutionary convergence in plant form: arid climate Avoiding erroneous inferences caused by homoplasy… 1. Examine traits at more than one developmental stage. e.g., As adults, limpets and barnacles are superficially similar, but larval anatomy reveals true phylogenetic relationship barnacle (crustacean) limpet (mollusk) Milkweed family Spurge family Cactus family (Asclepiadaceae) (Euphorbiaceae) (Cactaceae) crab (crustacean) Mollusks Traits evolving under strong natural selection may be most likely to show evolutionary convergence crabs barnacle (plus convergence to look/smell like carrion) Avoiding erroneous inferences caused by homoplasy… 2. Look at many traits (=characters). Phylogenetic signal from homologous traits will drown out ‘noise’ from homoplasy Looking at multiple characters… Methodology for distinguishing homology from homoplasy: The key: Synapomorphies reflecting homology require only 1 change on a phylogeny, whereas homoplasies require 2 or more changes. e.g., for a known phylogeny: if nucleotide position 1: G to A is a synapomorphy reflecting homology Fish eye Octopus eye Sp. 1 Sp. 2 A A Sp. 3 G Sp. 4 G Outgroup G G to A 5 Methodology for distinguishing homology from homoplasy: Synapomorphies reflecting homology require only 1 change on a phylogeny, whereas homoplasies require 2 or more changes. So… If we look at many characters, the phylogeny that requires the fewest number of changes (=steps) is the most probable true phylogeny. = ‘Maximum parsimony’ criterion (principle of Occam’s razor) e.g., for a known phylogeny: if nucleotide position 1: G to A is a synapomorphy reflecting homology and position 2: T to C is a homoplasy Sp. 1 AT Sp. 2 Sp. 3 AC GT T to C Sp. 4 GC e.g., for three species (ingroup), 3 possible trees: Outgroup GT outgroups (=OG) T to C 1 G to A 2 3 4 5 1 3 2 (4 OG 5) OG 2 3 1 (4 5) Homoplasies require more steps (changes) on a tree than homologous characters . If we’re looking at 7 variable nucleotide positions (a-g)… Tree 1 requires 8 changes (steps): tree length, L= 8 Tree 2 requires 9 changes (steps): tree length = 9 Consistency Index (CI): min. no. possible steps/ number of steps on tree Here, CI = 7/8 = 0.88 (for CI= 1, no homoplasy) Consistency Index, CI = 7/9 = 0.78 6 Tree 3 requires 10 changes (steps): tree length = 10 So tree 1 is most parsimonious (8 steps, CI = 0.88) Consistency Index, CI = 7/10 = 0.70 Grouping whales with fish based on the presence of dorsal fins is less parsimonious than true phylogeny: CI = 9/17 = 0.53 CI = 9/10 = 0.90 7