Restriction digest and gel electrophoresis
advertisement

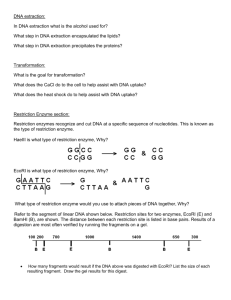
Restriction Digest 1 2 Restriction Enzymes/Endonucleases • Restriction Enzyme/Endonuclease - recognizes short DNA sequences and cleaves double stranded DNA at a particular sequence. – The recognition sequences are generally 4 to 6 nucleotides in length. – Called an ‘endonuclease’ because it cleaves within a nucleotide sequence. • Found naturally in bacteria. – Used by bacteria to degrade foreign DNA (bacteriophage). – Bacteria protect their own chromosomal DNA by methylating the nucleotides within the recognition sequence. Restriction Enzymes/Endonucleases • Found naturally in bacteria. – Used by bacteria to degrade foreign DNA (bacteriophage). – Bacteria protect their own chromosomal DNA by methylating the nucleotides within the recognition sequence. 3 Restriction Enzymes/Endonucleases • EcoRI from Escherichia coli • BamHI from Bacillus amyloliqueraciens • PvuI and PvuII are different enzymes from same strain. • Originally purified by individual labs, Nathans, Smith • Now supplied by companies - GE, NEB, Promega, BRL Restriction Enzymes/Endonucleases • “flush” or “blunt” ends – DraI cleavage generates blunt ends: 5´ T-T-T≠A-A-A 3´ 3´ A-A-A≠T-T-T 5´ • “sticky ends.” – KpnI cleavage generates cohesive 3´ overhanging ends: 5´ G-G-T-A-C≠C 3´ 3´ C≠C-A-T-G-G 5´ 4 Restriction Enzymes/Endonucleases Restriction Enzymes/Endonucleases • Degenerate (Recognizes multiple sequences): • AvaII GGWCC: GGTCC, GGACC • AvaI CPyCGPuG Py stands for pyrimidine- T or C Pu stands for purine - A or G CTCGAG CTCGGG CCCGAG CCCGGG • DdeI CTNAG: CTAAG, CTGAG, CTCAG, CTTAG • BbsI cleaves GAAGACNN CTTCTGNNNNNN 5 Restriction Enzymes/Endonucleases Restriction Enzymes/Endonucleases • It is possible that two or more restriction endonucleases recognizing different sequences may generate compatible ends. – BgIII and BamHI recognize two different sequences but produce compatible ends. BamHI BglII 6 Restriction Enzymes/Endonucleases • Mg+2 or Ca+2 is requirement for enzyme activity. • A buffer is necessary in order to maintain the optimal pH for the enzyme function. • When more than one restriction endonuclease is used in a reaction a buffer compatible with both enzymes must be used. Restriction Enzymes/Endonucleases • How much enzyme to use – Unit = Amount of enzyme required to digest 1µg of DNA in 1hr under optimal conditions – Generally 5-10 Units are used per digestion • How much DNA to use – .1-5 µg • Use the correct buffer • Incubation time (1-2 hrs) • Incubation temperature (generally 37°C) 7 Restriction Enzymes/Endonucleases • As a general rule of thumb the concentration of DNA obtained from the mini prep is approximately .1µg/µl. • We will digest 10µl of DNA from the mini prep. 10µl of mini prep DNA is 1 µg of DNA. • A Unit of enzyme is the amount of enzyme required to digest 1µg of DNA in 1hr under optimal conditions and 1µl of enzyme is about 10 Units. • Will this be enough to digest our concentration of DNA? • What is the minimum amount of enzyme required for each sample for our concentration of DNA? How many Units of enzyme is this? 8 • Restriction enzymes can be used to determine the size of the Wolffia cDNA insert. 9 Important! • Enzymes should be stored at -20°C. • While in use, enzymes should be carefully maintained on ice. • Be careful to avoid contaminating enzyme solutions. • Add the enzyme last just in case you make any mistakes with the other reagents! Today’s Experiment • Digest the purified plasmid DNA in order to determine the size of the insert • Use the restriction endonuclease PvuII to digest the plasmid • Complete a digest for each mini prep. 10 Today’s Experiment • Master Mix – Label a microfuge tube as “5-Rxns. Digest Mix.” In this tube prepare a reaction digestion mixture. – Notice that there will be more mix than you will need (for 5 rather than 4 reactions). This is done to allow for pipetting error so you don’t run out of mix. Today’s Experiment • • Mix the ingredients of the “5-Rxns Digest Mix” by gently pipetting the mix up and down a few times until mixed. (Do Not Vortex!) Aliquot 10 ul of the “5Rxns. Digest Mix” to each of four fresh, appropriately labeled microcentrifuge tubes, then add 10 ul of each plasmid DNA to obtain a final reaction volume of 20 ul. 11 Today’s Experiment • Gently mix each digest by tapping the tube. • Place your digest tubes in a sample rack and incubate the rack of samples in the 37ºC water bath for 5 to 10 minutes. • When your digests are done incubating, store your samples in your –20ºC freezer box until you are ready to load them onto an agarose gel. Today’s Experiment • REMEMBER: KEEP THE RESTRICTION ENZYME ON ICE AT ALL TIMES! 12 Theoretical PvuII digests of plasmids p. 2B-10 Gel Electrophoresis • Uncut DNA is used as a control. • Three bands appear in the uncut lane. • Open circle/Nicked DNA - Nicked plasmid DNA runs about 2x slower than linear DNA • Linear DNA - Some of the plasmid DNA gets cut open during the mini prep and runs to its true size. • Supercoil - Uncut plasmid DNA runs faster than linear or nicked DNA 4-4 13 A real Digest gel of clones from the Duckweed cDNA library p. 2B-11 1.Did the gel run OK? A) Yes B) No C) Can’t tell 14 2.Is there DNA in your minipreps? Are there bands in the uncut sample? A) Yes B) No C) Can’t tell p. 2B-11 3.Is there the same level of DNA in your minipreps? A) Yes B) No C) Can’t tell p. 2B-11 15 4. Did your DNA cut? Does your digested sample look different than your uncut? A) Yes B) No C) Can’t tell p. 2-18 5. Does your digest look complete? Are the intensities of the bands proportional to the size? p. 2B-12 16 Bands Generated from Partial Digests p. 2-19 6. Is there a PvuII site in the insert? Are there multiple bands? p. 2B-12 17 6. Is there a PvuII site in the insert? Are there multiple bands? p. 2B-13 7. What is the size of the insert? 2.9 Kb 700 bp - 700 bp = 0 bp p. 2B-13 18 8. Do the results from the digest and the PCR agree? 700 bp - 700 bp = 0 bp 200 bp - 200 bp = 0 bp p. 2B-14 8. Do the results from the 20JM1.10 digest and the PCR agree? 1100 bp - 700 bp = 400 bp 600 bp - 200 bp = 400 bp p. 2B-14 19 8. Do the results from the 20JM2.10 digest and the PCR agree? 2200 bp - 700 bp = 1500 bp 1700 bp - 200 bp = 1500 bp p. 2B-14 8. How do we figure out the insert size from the 20JM4.10 digest? 2900 bp 1200 bp 400 bp 1200 bp + 400 bp700 bp = 900 bp p. 2B-14 20 8. Can we map the position of the PvuII site? A) 2900 bp 1200 bp B) 400 bp p. 2B-14 8. Do the results from the 20JM4.10 digest and the PCR agree? 1100 bp - 200 bp = 900 bp 1200 bp +400 bp700 bp = 900 bp p. 2B-14 21 Home Work!! Fill in the rest of the table for Chapter 2B Problem Set Protocol • Wear gloves! There is ethidium bromide in the buffer! • Make 70ml of a 1% agarose in 1x TBE. Use an Erlenmeyer flask for this solution. 22 Protocol • Use a Kim wipe or a piece of paper towel as a stopper for the flask and heat the solution in the microwave for ~2 minutes. Protocol • Make sure that the agarose completely dissolves. • Allow to cool to ~60ºC. (This should take a couple of minutes. Estimate by touch) 23 Protocol • Pour the solution onto the casting tray. • Add the comb to the liquid agarose. • Allow to set. (~30 minutes) Protocol • Carefully remove the comb from the gel • Place the gel into the electrophoresis chamber in the proper orientation so that the negatively charged DNA runs toward the anode (+) 24 Protocol • Add enough 1x TBE to cover the gel Protocol • Obtain your restriction digests from the freezer and add 2 µl of 10X Gel Loading Dye Mix to each of your four restriction digest tubes. 25 Protocol • Obtain uncut mini prep DNA from the freezer Protocol • Arrange the samples in the order you will load the gel. • Record this information in your lab notebook. 26 Protocol • Load the DNA samples (all are approx. 20 µl) into wells using a P-20 pipetman set at 20 µl. In addition, load one lane of 10 µl of premade DNA size standards known as 1Kb Plus DNA Ladder. Protocol 27 Protocol • Attach the lid to the gel box. • Plug the leads into the power supply and set it to 100V. • Turn on the power supply and press the run button. Protocol • After you have turned on the gel, make sure that bubbles are coming from the wire electrodes, indicating that there is current in the gel. 28 Protocol • Turn off the power supply after the tracking dye runs about 2/3 of the way down the gel. Protocol • Remove the gel from the chamber (Be sure to keep track of the gel’s orientation). • Place the gel on the UV light box. • Take a picture. 29 Protocol • Tape the picture into your lab notebook. • Label the gel and analyze the bands in each lane. • Record your results. Protocol • Download the RDG PowerPoint template from the website. • Crop and mark up your gel • Save your gel using the proper naming format • Upload your marked up gel to the Sakai drop box. 30