AMS394.01 Practice Midterm Fall 2015 Name: ID: Signature:
advertisement

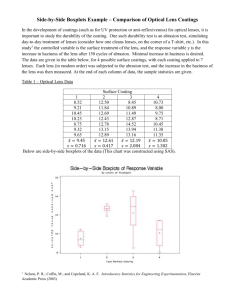
AMS394.01 Practice Midterm Fall 2015 Name: ____________________________ ID: ___________________ Signature: ___________________ Instruction: This is an open book exam. However no communication is allowed between students. Please provide complete solutions for full credit. Good luck! 1. We want to test the relative durability of 4 different surface coatings for optical lenses. The durability test involves subjecting a coated lens to 150 cycles of abrasion. The response variable is a measure of the increase in lens haziness. Please write up the SAS code, and the R code to do the following. In addition, please provide the out and summary of your tests/plots using one of these two programs: (1) We are testing the two hypotheses H0: 1 = 2 = 3= 4 vs. Ha: At least one of the means differs from the others. (2) Please include the follow-up tests for detecting specific differences among the means. (3) Please also include the side-by-side boxplot to check for homogeneity of variances, and, a residual plot. (4) Please conduct a usual t-test to compare the mean haziness between coatings 1 and 2. Solution: data one; input coating haziness; label coating = "Lens Surface Coating" haziness = "Lens Haziness after Abrasion"; datalines; 1 8.52 1 9.21 1 10.45 1 10.23 1 8.75 1 9.32 1 9.65 2 12.50 2 11.84 2 12.69 2 12.43 2 12.78 2 13.15 2 12.89 3 8.45 3 10.89 3 11.49 3 12.87 3 14.52 3 13.94 3 13.16 1 4 10.73 4 8.00 4 9.75 4 8.71 4 10.45 4 11.38 4 11.35 ; Run; proc boxplot data=one; plot haziness*coating; title "Side-by-Side Boxplots of Response Variable"; title2 "by Levels of Treatment"; Run; Proc glm data=one; class coating; model haziness = coating; lsmeans coating /out=outmns; means coating / cldiff bon; output out=resout p=preds rstudent=exstdres; title "Analysis of Variance for Optical Lens Surface Coatings"; title2 "With Follow-Up Tests"; Run; Quit; title 'Profile Plot'; symbol i=j; proc gplot data=outmns; where coating ne .; plot lsmean*coating; run; quit; goptions reset=all; title 'Residual Plot'; proc gplot data=resout; plot exstdres*preds; run; quit; data two; input coating haziness; label coating = "Lens Surface Coating" 2 haziness = "Lens Haziness after Abrasion"; datalines; 1 8.52 1 9.21 1 10.45 1 10.23 1 8.75 1 9.32 1 9.65 2 12.50 2 11.84 2 12.69 2 12.43 2 12.78 2 13.15 2 12.89 ; Run; Proc univariate data=two normal; Class coating; Var haziness; Title ‘check for normality’; Run; Proc ttest data=two; Class coating; Var haziness; Title ‘Independent samples t-test’; Run; 3 Selected output and summary: (1) The GLM Procedure Dependent Variable: haziness Lens Haziness after Abrasion Sum of Source DF Squares Mean Square F Value Pr > F Model 3 51.06744286 17.02248095 10.12 0.0002 Error 24 40.35205714 1.68133571 Corrected Total 27 91.41950000 Summary: we reject the ANOVA null hypothesis. (2) The GLM Procedure Bonferroni (Dunn) t Tests for haziness NOTE: This test controls the Type I experimentwise error rate, but it generally has a higher Type II error rate than Tukey's for all pairwise comparisons. Alpha 0.05 Error Degrees of Freedom Error Mean Square 24 1.681336 Critical Value of t 2.87509 Minimum Significant Difference 1.9927 Comparisons significant at the 0.05 level are indicated by ***. Difference coating Comparison Between Simultaneous 95% Means Confidence Limits 2 - 3 0.4229 -1.5699 2.4156 2 - 4 2.5586 0.5659 4.5513 *** 2 - 1 3.1643 1.1716 5.1570 *** 3 - 2 -0.4229 -2.4156 3 - 4 2.1357 0.1430 4.1284 *** 3 - 1 2.7414 0.7487 4.7341 *** 4 - 2 -2.5586 -4.5513 1.5699 -0.5659 *** 4 4 - 3 -2.1357 -4.1284 -0.1430 *** 4 - 1 0.6057 -1.3870 2.5984 1 - 2 -3.1643 -5.1570 -1.1716 *** 1 - 3 -2.7414 -4.7341 -0.7487 *** 1 - 4 -0.6057 -2.5984 1.3870 Summary: the pairwise comparisons show that coatings 1/2, 1/3, 2/4, 3/4 are significantly different at the familywise error rate of 0.05. Note, although we used the Bonferroni method here as an example, the Tukey method is less conservative and in general, better. (3) Side-by-Side Boxplots of Response Variable by Levels of Treatment Lens Haziness after Abrasion 16 14 12 10 8 1 2 3 4 Lens Surface Coating 5 Residual Plot exstdres 3 2 1 0 -1 -2 -3 -4 9 10 11 12 13 preds Summary: The box-plots make us worry about the equal variance assumptions. The residual plot shows some concern of unequal variance too. (4) The UNIVARIATE Procedure Variable: haziness (Lens Haziness after Abrasion) coating = 1 Tests for Normality Test --Statistic--- Shapiro-Wilk W 0.953597 -----p Value------ Pr < W 0.7623 Kolmogorov-Smirnov D 0.148529 Pr > D >0.1500 Cramer-von Mises W-Sq 0.027158 Pr > W-Sq >0.2500 Anderson-Darling A-Sq 0.196002 Pr > A-Sq >0.2500 The UNIVARIATE Procedure Variable: haziness (Lens Haziness after Abrasion) coating = 2 Tests for Normality Test Shapiro-Wilk Kolmogorov-Smirnov --Statistic--- W D 0.949828 0.188846 -----p Value------ Pr < W 0.7281 Pr > D >0.1500 6 Cramer-von Mises W-Sq 0.036567 Pr > W-Sq >0.2500 Anderson-Darling A-Sq 0.251295 Pr > A-Sq >0.2500 Summary: The Shapiro-Wilk test shows that both samples are normal and thus we can continue with the independent samples t-test. The TTEST Procedure Variable: coating haziness N (Lens Haziness after Abrasion) Mean Std Dev Std Err Minimum Maximum 1 7 9.4471 0.7162 0.2707 8.5200 10.4500 2 7 12.6114 0.4169 0.1576 11.8400 13.1500 Diff (1-2) coating -3.1643 Method 0.5860 Mean 0.3132 95% CL Mean Std Dev 95% CL Std Dev 1 9.4471 8.7848 10.1095 0.7162 0.4615 1.5771 2 12.6114 12.2259 12.9970 0.4169 0.2686 0.9180 Diff (1-2) Pooled -3.1643 Diff (1-2) Satterthwaite -3.1643 Method Variances Pooled Equal Satterthwaite Unequal -3.8467 -3.8657 -2.4818 0.5860 0.4202 0.9673 -2.4629 DF t Value Pr > |t| 12 -10.10 <.0001 9.6468 -10.10 <.0001 Equality of Variances Method Folded F Num DF 6 Den DF 6 F Value 2.95 Pr > F 0.2135 Summary: The F-test shows that the variances can be considered equal. Therefore we adopted the pooled-variance t-test and found significant mean differences (in terms of haziness of lenses) between coatings 1 and 2. 2. The following SAS data step inputs a two-way ANOVA data set examining the relationship between crop density, amount of fertilizers, and crop yield. Please write up the SAS code, and the R code to do the following. In addition, please provide the out and summary of your tests/plots using one of these two programs: 7 (1) We are testing the ANOVA hypotheses of (a) no interaction, (b) density main effect, and (c) fertilizer main effect. (2) Please include the follow-up tests for detecting specific differences among the means. (3) Please also include the side-by-side boxplot to check for homogeneity of variances, and, a residual plot. PROC FORMAT; VALUE den 1='regular' 2='thick'; VALUE fert 1='low' 2='medium' 3='high'; RUN; DATA soybean(DROP=rep); FORMAT density den. fertilizer fert.; DO fertilizer = 1 TO 3; DO density = 1 TO 2; DO rep = 1 TO 4; INPUT yield @@; OUTPUT; END; END; END; DATALINES; 37.5 36.5 38.6 36.5 37.4 35.0 38.1 36.5 48.1 48.3 48.6 46.4 36.7 36.4 39.3 37.5 48.5 46.1 49.1 48.2 45.7 45.7 48.0 46.4 ; Run; Proc sort data=soybean; By fertilizer; Run; proc boxplot data=soybean; plot yield*fertilizer; title "Side-by-Side Boxplot of Response Variable"; title2 "by Levels of fertilizer"; Run; Proc sort data=soybean; By density; Run; proc boxplot data=soybean; plot yield*density; 8 title "Side-by-Side Boxplot of Response Variable"; title2 "by Levels of density"; Run; TITLE3 'Tests for Interaction & Main Effects'; PROC GLM DATA=soybean ORDER=INTERNAL; CLASS density fertilizer; MODEL yield = density | fertilizer; lsmeans density fertilizer density*fertilizer /out=outmns; means density fertilizer /cldiff bon; output out=resout p=preds rstudent=exstdres; RUN; Quit; title 'Profile/Interaction Plots'; symbol i=j; proc gplot data=outmns; where fertilizer ne . and density ne .; plot lsmean*density=fertilizer; plot lsmean*fertilizer=density; run; quit; goptions reset=all; *resets PROC GPLOT options; title 'Residual Plot'; proc gplot data=resout; plot exstdres*preds; run; quit; 9 Selected output and summary: (1) Source DF Type III SS Mean Square F Value Pr > F density 1 102.9204167 102.9204167 74.01 <.0001 fertilizer 2 417.7733333 208.8866667 150.20 <.0001 42.27 <.0001 density*fertilizer 2 117.5633333 58.7816667 Summary: we see significant interaction and main effects. (2) Bonferroni (Dunn) t Tests for yield NOTE: This test controls the Type I experimentwise error rate, but it generally has a higher Type II error rate than Tukey's for all pairwise comparisons. Alpha 0.05 Error Degrees of Freedom 18 Error Mean Square 1.390694 Critical Value of t 2.10092 Minimum Significant Difference 1.0115 Comparisons significant at the 0.05 level are indicated by ***. Difference density Between Simultaneous 95% Comparison Means Confidence Limits regular - thick 4.1417 3.1302 -4.1417 -5.1531 thick - regular 5.1531 *** -3.1302 *** Bonferroni (Dunn) t Tests for yield NOTE: This test controls the Type I experimentwise error rate, but it generally has a higher Type II error rate than Tukey's for all pairwise comparisons. Alpha Error Degrees of Freedom Error Mean Square Critical Value of t 0.05 18 1.390694 2.63914 10 Minimum Significant Difference 1.5561 Comparisons significant at the 0.05 level are indicated by ***. Difference fertilizer Between Simultaneous 95% Comparison Means Confidence Limits high - medium 4.5500 2.9939 high - low 10.2000 8.6439 11.7561 *** medium - high -4.5500 -6.1061 -2.9939 *** medium - low 5.6500 4.0939 -10.2000 -11.7561 -8.6439 *** -5.6500 -7.2061 -4.0939 *** low - high low - medium 6.1061 7.2061 *** *** Summary: the pairwise comparisons show that all pairs are significantly different from each other in means. (3) Side-by-Side Boxplot of Response Variable by Levels of fertilizer 50.0 47.5 yield 45.0 42.5 40.0 37.5 35.0 low medium high fertilizer 11 Side-by-Side Boxplot of Response Variable by Levels of density 50.0 47.5 yield 45.0 42.5 40.0 37.5 35.0 regular thick density Residual Plot exstdres 2 1 0 -1 -2 36 37 38 39 40 41 42 43 44 45 46 47 48 preds Summary: The box-plots make us worry about the equal variance assumptions for different fertilizers, but no worries for different density levels. The residual plot seems okay. 12 3. The following dataset examines the relationship between time to headache relief, and the three brands of pain killers. Please use the REGRESSION procedures in SAS and R to analyze this data set. (1) Please write down the program for both SAS and R, and use one of these two programs to analyze the data. (2) Please include necessary plots and analyses to verify the underlying model assumptions. (3) Please include your output and summary of results. Data three; Input BRAND RELIEF; Dummy1= 0; Dummy2= 0; If brand=1 then dummy1=1; If brand=2 then dummy2=1; Datalines; 1 24.5 1 23.5 1 26.4 1 27.1 1 29.9 2 28.4 2 34.2 2 29.5 2 32.2 2 30.1 3 26.1 3 28.3 3 24.3 3 26.2 3 27.8 ; Run; Proc print data=three; Run; proc boxplot data=three; plot relief*brand; title "Side-by-Side Boxplots of Response Variable"; title2 "by brands of Treatment"; Run; Proc glm data=three; 13 class brand; model relief = brand; lsmeans brand /out=outmns; means brand / cldiff bon; output out=resout p=preds rstudent=exstdres; title "Analysis of Variance for Pain Relief by Drug Brands"; title2 "With Follow-Up Tests"; Run; Quit; Proc reg data=three; model relief = dummy1 dummy2; Run; Quit; title 'Profile Plot'; symbol i=j; proc gplot data=outmns; where brand ne .; plot lsmean*brand; run; quit; goptions reset=all; title 'Residual Plot'; proc gplot data=resout; plot exstdres*preds; run; quit; 14 Selected output and summary: Dear students, the only difference of what is required in this problem versus that in Problem 1, is that I need you to write down the general linear model. This can be accomplished by you setting up the dummy variables and then run the regression with the dummy variables directly. There will be other approaches but we are showing the easiest one here. So to save time, I will only show this different part. Obs BRAND RELIEF Dummy1 Dummy2 1 1 24.5 1 0 2 1 23.5 1 0 3 1 26.4 1 0 4 1 27.1 1 0 5 1 29.9 1 0 6 2 28.4 0 1 7 2 34.2 0 1 8 2 29.5 0 1 9 2 32.2 0 1 10 2 30.1 0 1 11 3 26.1 0 0 12 3 28.3 0 0 13 3 24.3 0 0 14 3 26.2 0 0 15 3 27.8 0 0 Parameter Estimates Parameter Standard Variable DF Estimate Error t Value Pr > |t| Intercept 1 26.54000 0.96720 27.44 <.0001 Dummy1 1 -0.26000 1.36782 -0.19 0.8524 Dummy2 1 4.34000 1.36782 3.17 0.0080 Summary: Here you see the dataset with the two dummy variables. The estimated general linear model is: Yˆ 26.54 0.26* dummy1 4.34* dummy 2 15