nature répétitive du génome des eucaryotes
advertisement
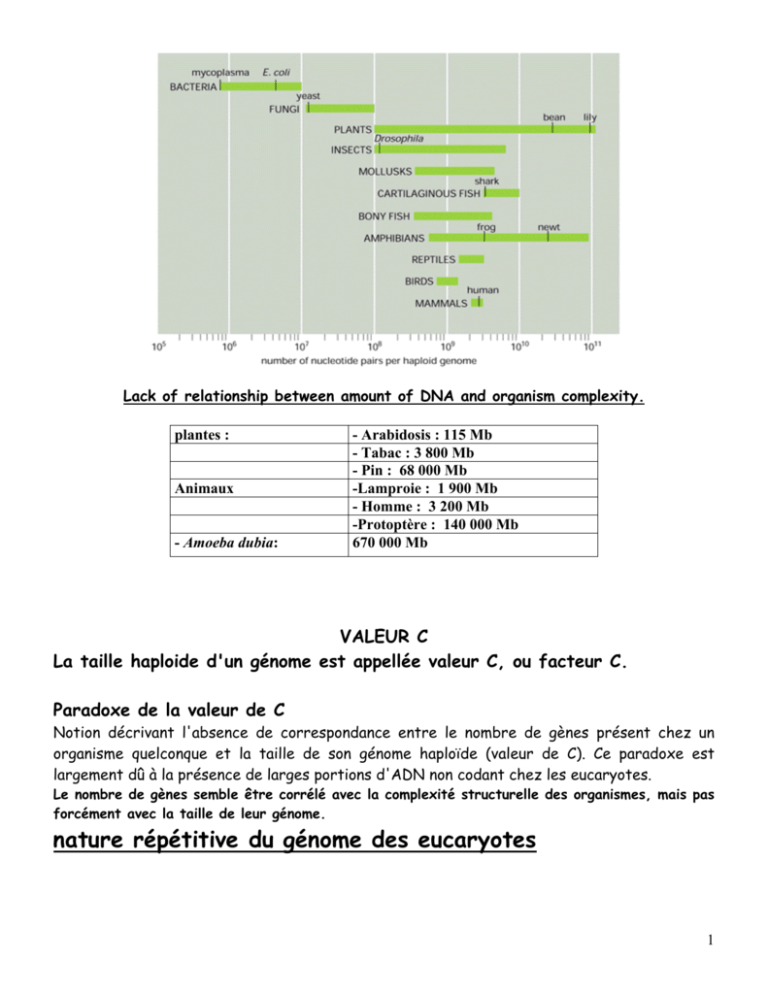
Lack of relationship between amount of DNA and organism complexity. plantes : Animaux - Amoeba dubia: - Arabidosis : 115 Mb - Tabac : 3 800 Mb - Pin : 68 000 Mb -Lamproie : 1 900 Mb - Homme : 3 200 Mb -Protoptère : 140 000 Mb 670 000 Mb VALEUR C La taille haploide d'un génome est appellée valeur C, ou facteur C. Paradoxe de la valeur de C Notion décrivant l'absence de correspondance entre le nombre de gènes présent chez un organisme quelconque et la taille de son génome haploïde (valeur de C). Ce paradoxe est largement dû à la présence de larges portions d'ADN non codant chez les eucaryotes. Le nombre de gènes semble être corrélé avec la complexité structurelle des organismes, mais pas forcément avec la taille de leur génome. nature répétitive du génome des eucaryotes 1 The genome, transcriptome and proteome. 2 Gene Size mRNA Size Number of Introns β-Globin 1.5 0.6 2 Insulin 1.7 0.4 2 Protein kinase C 11 1.4 7 Albumin 25 2.1 14 Catalase 34 1.6 12 LDL receptor 45 5.5 17 Factor VIII 186 9 25 Thyroglobulin 300 8.7 36 more than 2000 17 more than 50 Dystrophin The Size of Some Human Genes in Thousands of Nucleotides The size specified here for a gene includes both its transcribed portien and nearby regulatory DNA sequences. Summary of the steps leading from gene to protein in eucaryotes and bacteria. 3 Six steps at which eucaryotic gene expression can be controlled. Many steps in the pathway from RNA to protein are regulated by cells to control gene expression. Most genes are thought to be regulated at multiple levels, although control of the initiation of transcription (transcriptional control, step 1) usually predominates. Some genes, however, are transcribed at a constant level and turned on and off solely by post-transcriptional regulatory processes, steps 2-5. These processes include (A) alternative RNA splice-site selection, (B) control of 3′-end formation by cleavage and poly-A addition, (C) RNA editing, (D) control of transport from the nucleus to the cytosol, (E) localization of mRNAs to particular parts of the cell, (F) control of translation initiation, and (G) regulated mRNA degradation. Most of these control processes require the recognition of specific sequences or structures in the RNA molecule being regulated. This recognition is accomplished by either a regulatory protein or a regulatory RNA molecule. Step 6, the regulation of protein activity, includes reversible activation or inactivation by protein phosphorylation as well as irreversible inactivation by proteolytic degradation. INTERACTIONS PROTEINES/ADN ; PROTEINES/ARN ; PROTEINES/ PROTEINES ; ARN/ARN 4 A Whole Bunch of Control Points for Messenger RNAs 5 The RNA content of a cell. Principal Types of RNAs Produced in Cells TYPE OF RNA FUNCTION mRNAs messenger RNAs, code for proteins rRNAs ribosomal RNAs, form the basic structure of the ribosome and catalyze protein synthesis tRNAs transfer RNAs, central to protein synthesis as adaptors between mRNA and amino acids snRNAs small nuclear RNAs, function in a variety of nuclear processes, including the splicing of pre-mRNA snoRNAs small nucleolar RNAs, used to process and chemically modify rRNAs Other noncoding RNAs function in diverse cellular processes, including telomere synthesis, X-chromosome inactivation, the transport of proteins into the ER, miRNA: MicroRNA — putative translational regulatory gene family siRNA: Small interfering RNA — active molecules in RNA interference Non-coding RNAs seem to be particularly abundant in roles that require highly specific nucleic acid recognition without complex catalysis, such as in directing post-transcriptional regulation of gene expression or in guiding RNA modifications. 6 Schematic view of subnuclear structures A typical vertebrate nucleus has several Cajal bodies, which are proposed to be the sites where snRNPs and snoRNPs undergo their final modifications. Interchromatin granule clusters are proposed to be storage sites for fully mature snRNPs. A typical vertebrate nucleus has 20–50 interchromatin granule clusters. After their initial synthesis, snRNAs are exported from the nucleus, after which they undergo 5′ and 3′ end-processing and assemble with the seven common snRNP proteins (called Sm proteins). These complexes are reimported into the nucleus and the snRNPs undergo their final modification in Cajal bodies. In addition, the U6 snRNP requires chemical modification by snoRNAs in the nucleolus. The sites of active transcription and splicing (approximately 2000–3000 sites per vertebrate nucleus) correspond to the “perichromatin fibers” seen under the electron microscope. The Nucleus is not a Loose Bag of DNA and Enzymes: Localized Centers of Transcription and RNA Processing 7 The function of the nucleolus in ribosome synthesis. The 45S rRNA transcript is packaged in a large ribonucleoprotein particle containing many ribosomal proteins imported from the cytoplasm. While this particle remains in the nucleolus, selected pieces are discarded as it is processed into immature large and small ribosomal subunits. These two subunits are thought to attain their final functional form only as each is individually transported through the nuclear pores into the cytoplasm. 8 RNA POLYMERASES Only one major RNA polymerase activity can be extracted from E. coli cells and from most other bacteria. Three major RNA polymerases can be extracted from nuclei of cells of humans and other eukaryotes. Single RNA polymerases can be extracted from the mitochondria and one or two from chloroplasts. These enzymes vary in their sensitivity to inhibitors. Consists of 2α, 1β, 1β’ subunits and a σ factor ; σ70 is the common one. σ factor disassociates after 10 nucleotides. Eucaryotic RNA polymerase - large and complex, many subunits. 3 different polymerases - PolI, PolII, PolIII which transcribe different types of genes. RNA Polymerase I RNA product RNA Polymerase II -nuclear pre-messenger RNA, pre- mRNA, (also called heterogeneous -ribosomal RNA nuclear RNA, or hnRNA,in (rRNA) except the 5S older work); rRNA RNA Polymerase III -transfer RNA (tRNA); -5S rRNA; -other small RNAs -some small nuclear RNAs (snRNA) sensitivity to αamanitin resistant 50% inhibition at 0.02 mg/ml sensitivity to actinomycin D very sensitive slightly sensitive 50% inhibition at 20 mg/ml slightly sensitive 9 Sensitivity of the three RNA polymerases to α-amanitin Pol II is used to make mRNA: 12 subunits (10 - 220 kDa) 220 kDa subunit (largest) resembles E. coli β' Has a C-terminal domain (CTD) consisting of a repeated sequence that can be phosphorylated. CTD of RNA Pol II: Coupling of Transcription, Elongation and RNA Processing 10 Sequence of the human CTD. Self-alignment of human RNAP II largest subunit sequence from amino acids 1593-1970 (last residue before the stop codon). The “RNA factory” concept for eucaryotic RNA polymerase II. Not only does the polymerase transcribe DNA into RNA, but it also carries pre-mRNA-processing proteins on its tail, which are then transferred to the nascent RNA at the appropriate time. There are many RNA-processing enzymes, and not all travel with the polymerase. For RNA splicing, for example, only a few critical components are carried on the tail; once transferred to an RNA molecule, they serve as a nucleation site for the remaining components. The RNA-processing proteins first bind to the RNA polymerase tail when it is phosphorylated late in the process of transcription initiation. 11 Eucaryotic RNA processing • • • • y p p g Eukaryotic mRNA precursor is called pre-mRNA, or hnRNA (heterogeneous nuclear RNA) RNA processing: is the structural and chemical maturation of newly synthesised RNA molecules 4 types of processing take place on most pre-mRNAs: 1. 5'-processing = addition of a 5'-cap structure 2. 3'-processing = cleavage or cleavage and polyadenylation 3. Intron removal and splicing exons together (splicing and alternative splicing) 4. Infrequent methylation Some pre-mRNAs are modified by RNA editing to change the sequence of the transcript 12 Eukaryotic pre-mRNA 5'-processing RNA Capping Is the First Modification of Eucaryotic Pre-mRNAs 13 Capping of the 5′ end of nascent RNA transcripts with 7′-methylguanylate (m7G). The first two reactions are catalyzed by a capping enzyme that associates with the phosphorylated CTD of RNA polymerase II shortly after transcription initiation. Two different methyltransferases catalyze reactions 3 and 4. S-adenosylmethionine (S-AdoMet) is the source of the methyl (CH3) group for the two methylation steps; the guanylate (G) is methylated first, then the 2′ hydroxyl of the first one or two nucleotides (N) in the transcript. Functions of Cap: In the nucleus, the cap binds a protein complex called CBC (cap-binding complex), which helps the RNA to be properly processed and exported. 1) Protect mRNAs from degradation. The cap seems to protect the mRNA from attack by RNases that begin at the 5'-end of their substrates that cannot cleave triphosphate linkages. 2) Enhance transport of mRNAs from the nucleus into the cytoplasm. 3) Enhance the efficiency of splicing of mRNAs. 4) The 5′ methyl cap also has an important role in the translation of mRNAs in the cytosol. Eukaryotic mRNAs gain access to the ribosome for translation via the cap-binding protein that recognizes the cap. If there is no cap, the cap-binding protein cannot bind and the mRNA is very poorly translated. The presence of the cap stimulates translation of mRNA about 300-fold. 14 Structure of the 5′ methylated cap of eukaryotic mRNA The distinguishing chemical features are the 5′ - 5′ linkage of 7-methylguanylate to the initial nucleotide of the mRNA molecule and the methyl group on the 2′ hydroxyl of the ribose of the first nucleotide (base 1). Both these features occur in all animal cells and in cells of higher plants; yeasts lack the methyl group on base 1. The ribose of the second nucleotide (base 2) also is methylated in vertebrates. 15 Transcription Termination and Polyadenylation Eukaryotic transcripts have polyA tails. These arise due to cleavage and addition of AAAAs. The signal for cleavage and polyadenylation is AAUAAA. The addition of poly[A] (polyadenylation) to pre-mRNA is part of the events that end the transcription process. The mechanism involves three steps: 1) Cleavage of the RNA chain at a particular site; 2) Addition of the poly [A] tail to the 3' end of the pre-mRNA; and 3) Degradation of the remainder of the RNA transcript. There is a specific sequence that dictates where the endonuclease required for cleaveage and poly [A] polymerase will act. This is the polyadenylation signal, given by the sequence AAUAAA in the pre-mRNA transcript. Cleavage and polyadenylation take place 23 or 24 bases downstream from this signal at a GU rich region of the RNA transcript that is followed immediately by a U-rich area. This sequence is quite well conserved with variations from this sequence decreasing the efficiency with which polyadenylation takes place. Factors involved: 16 17 Function of Poly (A): Most mRNAs contain poly(A), except for histone mRNAs. 1) Poly (A) is thought to protect mRNA from degradation. 2) It seems that the poly(A) tail stimulates translation of mRNAs. One of the proteins that binds to a eukaryotic mRNA during translation is poly(A) binding protein I (PAB I). Binding to this protein seems to boost the efficiency with which a mRNA is translated. Poly (A)+ mRNA forms polysomes more successfully than poly (A)mRNA. The true function of the poly[A] tail still requires further study. In the case of histone genes, which are unique in producing mRNA that does not become polyadenylated, termination of transcription also involves 3′ cleavage of the primary transcript. This reaction is dependent on secondary structure in the RNA transcript, including a conserved upstream hairpin sequence and a short downstream sequence which base-pairs with a short sequence at the 5′ end of the U7 snRNA 18 Factors involved in cleavage of the histone mRNA precursor. SLBP1/HBP, stem-loop binding protein 1/hairpin-binding factor; HLF, heat-labile factor. Poly-A site choice Function: Contributes to protein diversity via alternative choice of cleavage site 19 RNA splicing Process in which intron sequences are excised from RNA transcripts in the nucleus during formation of messenger and other RNAs. Four major classes of introns have been distinguished based on their mechanisms of splicing: Group I Self-Splicing Introns: These introns are found primarily in nucleolar rRNA genes and in organelle genomes - both in mitochondrial DNA (mtDNA) and in chloroplast DNA (ctDNA). No additional proteins and no energy source is required for the splicing reaction. However, a free guanosine nucleoside is required as the catalytic agent in the mechanism. Group II Self-Splicing Introns: This type of intron is found primarily in mtDNA and in ctDNA. As with Group I introns, no additional proteins and no energy source is required for the reaction. The catalytic agent is an internal hydroxyl group within the intron. Nuclear mRNA Spliceosomal Introns: The mechanism of the splicing reaction in nuclear mRNA introns is similar to that of the Group II Introns, splicing of these introns requires the participation of a specific set of protein-RNA particles. Nuclear tRNA Enzymatically Spliced Introns: These introns also require the help of enzymes to catalyze their removal but the mechanism is completely different - being a cut and rejoin type of mechanism. Intron type Where found GU-AG introns Eukaryotic nuclear pre-mRNA AU-AC introns Eukaryotic nuclear pre-mRNA Group I Eukaryotic nuclear pre-rRNA, organelle RNAs, few bacterial RNAs Group II Organelle RNAs, some prokaryotic RNAs Pre-tRNA introns Eukaryotic nuclear pre-tRNA 20 Self-splicing introns: Group I and group II self-splicing introns are distinguished by their reaction mechanisms. In group I introns, the first step in splicing is cleavage of the 5′ splice site by reaction with a guanosine cofactor. The result is a linear intermediate with a G added to the 5′ end of the intron. In group II introns (as in pre-mRNA splicing), the first step is cleavage of the 5′ splice site by reaction with an A within the intron, forming a lariat-like intermediate. In both cases, the second step is simultaneous cleavage of the 3′ splice site and ligation of the exons. Consensus sequences at exon-intron boundaries Consensus sequences around 5′ and 3′ splice sites in vertebrate pre-mRNAs. The only nearly invariant bases are the (5′)GU and (3′)AG of the intron, although the flanking bases indicated are found at frequencies higher than expected based on a random distribution. A pyrimidine-rich region (light blue) near the 3′ end of the intron is found in most cases. The branch-point adenosine, also invariant, usually is 20 – 50 bases from the 3′ splice site. The central region of the intron, which may range from 40 bases to 50 kilobases in length, generally is unnecessary for splicing to occur. 21 Splicing of pre-mRNA: The splicing reaction proceeds in two steps. The first step involves cleavage at the 5′ splice site (SS) and joining of the 5′ end of the intron to an A within the intron (the branch point). This reaction yields a lariat-like intermediate, in which the intron forms a loop. The second step is cleavage at the 3′ splice site and simultaneous ligation of the exons, resulting in excision of the intron as a lariat-like structure. Step 1 involves a nucleophilic attack by the 2'-OH group of the branch point A on the phosphodiester bond of the 5' exon/intron boundary, displacing the 5' exon as the leaving group and giving the lariat structure shown in the middle. The second step is a nucleophilic attack of the 3' end of the 5' exon on the phosphodiester bond of the 3' intron/exon boundary, displacing the intron (as a lariat form) and sealing the two exons together. 22 Splicing of exons in pre-mRNA occurs via two transesterification reactions. In the first reaction, the ester bond between the 5′ phosphorus of the intron and the 3′ oxygen of exon 1 is exchanged for an ester bond with the 2′ oxygen of the branch-site A residue. In the second reaction, the ester bond between the 5′ phosphorus of exon 2 and the 3′ oxygen of the intron is exchanged for an ester bond with the 3′ oxygen of exon 1, releasing the intron as a lariat structure and joining the two exons. Arrows show where the activated hydroxyl oxygens react with phosphorus atoms. Biochemical analysis of nuclear extracts has revealed that splicing takes place in large complexes, called spliceosomes, composed of proteins and RNAs. 23 The RNA components of the spliceosome are five types of small nuclear RNAs (snRNAs) called U1, U2, U4, U5, and U6. These snRNAs, which range in size from approximately 90 to 200 nucleotides, are complexed with six to ten protein molecules to form small nuclear ribonucleoprotein particles (snRNPs ou snurps), which play central roles in the splicing process. The U1, U2, and U5 snRNPs each contain a single snRNA molecule, whereas U4 and U6 snRNAs are complexed to each other in a single snRNP. snRNAs snRNA Length (nts) U1 165 U2 185 U4 116 Masks the catalytic activity of U6 U5 145 Binds the 5’ splice site U6 106 Catalyzes splicing Function Binds 5’ splice site, then 3’ splice site Binds the branch site and forms part of the catalytic center The spliceosome: the most complex macromolecular machine in the cell? Timothy W. Nilsen BioEssays 25:1147–1149, 2003. Proteomic analyses of purified spliceosomes reveal that the spliceosome is composed of as many as 300 distinct proteins and five RNAs, making it among the most complex macromolecular machines known. The spliceosome contains five small nuclear ribonucleoproteins that assemble onto the intron. The Early (E) complex contains the U1 snRNP bound to the 5’ splice site. Each element of the 3’ splice site is bound by a specific protein, the branch point by SF1 (BBP), the polypyrimidine tract by U2AF 65, and the AG dinucleotide by U2AF 35. This complex also apparently contains the U2 snRNP not yet bound to the branch point. The A complex forms when U2 engages the branch point via RNA/RNA base-pairing. This complex is joined by the U4/5/6 Tri-snRNP to form the B complex. The B complex is then extensively rearranged to form the catalytic C complex. During this rearrangement the interactions of the U1 and U4 snRNPs are lost and the U6 snRNP is brought into contact with the 5’ splice site. 24 Dynamic changes in spliceosome composition direct premRNA splicing 25 A proposed model for the mechanism of splicing. 26 The spliceosomal splicing cycle. The splicing snRNPs (U1, U2, U4, U5, and U6) associate with the pre-mRNA and with each other in an ordered sequence to form the spliceosome. This large ribonucleoprotein complex then catalyzes the two transesterification reactions that result in splicing of the exons and excision of the intron as a lariat structure. Although ATP hydrolysis is not required for the transesterification reactions, it is thought to provide the energy necessary for rearrangements of the spliceosome structure that occur during the cycle. Note that the snRNP proteins in the spliceosome are distinct from the hnRNP proteins. In higher eukaryotes, the association of U2 snRNP with pre-mRNA is assisted by an hnRNP protein called U2AF, which binds to the pyrimidine-rich region near the 3′ splice site. U2AF also probably interacts with other proteins required for splicing through a domain containing repeats of the dipeptide serine-arginine (the SR motif). The branch-point A in pre-mRNA is indicated in boldface. 27 Spliceosome and ATP Æ RNA-RNA Rearrangements - II rearrangement RNA helicases facilitate these rearrangements. Less understood are the earliest events that initiate splicing, in which the boundaries of the intron that must be removed are identified. The absolute accuracy of this molecular recognition is critical to gene function and must depend on signals comprised of sequence and/or structural elements in the RNA substrate. 28 ALTERNATIVE SPLICING The Definition of a Gene has had to be Modified Since the Discovery of Alternative RNA Splicing: generates variable segments within mRNAs. Alternative promoters: Selection of one of multiple first exons results in variability at the 5' terminus of the mRNA (1). Alternative splicing of internal exons: Alternative splicing patterns for internal exons include cassette (2), alternative 5' splice sites (3), alternative 3' splice sites (4), intron retention (5), and mutually exclusive (6). The effects on coding potential are an in-frame insertion or deletion, a reading-frame shift, or introduction of a stop codon. Introduction of a premature termination codon into an mRNA by alternative splicing can be a mechanism to down-regulate expression of a gene. Alternative terminal exons: The 3' end of an mRNA is determined by a directed cleavage event followed by addition of the poly(A) tail (Proudfoot et al. 2002). Selection of one of multiple terminal exons (7) results from a competition between cleavage at the upstream poly(A) site or splicing to the downstream 3' splice site. There are also examples of competition between a 5' splice site and a poly(A) site within an upstream terminal exon (8). Variability at the 3' end of the mRNA produces either proteins with different C termini or mRNAs with different 3'-UTRs. 29 Patterns of alternative splicing. Constitutive sequences present in all final mRNAs are gray boxes. Alternative RNA segments that may or may not be included in the mRNA are hatched boxes. (A) A cassette exon can be either included in the mRNA or excluded. (B) Mutually exclusive exons occur when two or more adjacent cassette exons are spliced such that only one exon in the group is included at a time. (C, D) Alternative 5’ and 3’ splice sites allow the lengthening or shortening of a particular exon. (E, F) Alternative promoters and alternative poly(A) sites switch the 5’ or 3’ most exons of a transcript. (G) A retained intron can be excised from the pre-mRNA or can be retained in the translated mRNA. (H) A single pre-mRNA can exhibit multiple sites of alternative splicing using different patterns of inclusion. These are often used in a combinatorial manner to produce many different final mRNAs. Two types of splicing errors. 30 Genetics, Vol. 159, 599-608, October 2001 : Alternative Splicing of the Drosophila Dscam Pre-mRNA Is Both Temporally and Spatially Regulated . Alicia M. Celotto and Brenton R. Graveley Axon guidance receptors Æ direct growth cones to appropriate targets in developing nervous system It was estimated that at least 50% of all human genes are subject to alternative splicing. Approximately 15% of human genetic diseases and developmental defects have been correlated with disruptions of alternative splicing control in particular genes. The regulation of alternative splicing is a complex process involving multiple steps. Pre-mRNA sequences important for splicing. 31 Consensus mammalian 5’ splice site (5’ SS), branch site (BS; indicated by a black circle), polypyrimidine tract (PPT) and 3’ splice site (3’SS) sequences are shown. ESE, exonic splicing enhancer; R, purine; Y, pyrimidine. The exon definition hypothesis: role of SR proteins According to one proposal, SR proteins bind to each exon sequence in the pre-mRNA and thereby help to guide the snRNPs to the proper intron/exon boundaries. This demarcation of exons by the SR proteins occurs co-transcriptionally, beginning at the CBC (cap-binding complex) at the 5′ end. As indicated, the intron sequences in the pre-mRNA, which can be extremely long, are packaged into hnRNP (heterogeneous nuclear ribonucleoprotein) complexes that compact them into more manageable structures and perhaps mask cryptic splice sites. Each hnRNP complex forms a particle approximately twice the diameter of a nucleosome, and the core is composed of a set of at least eight different proteins. It has been proposed that hnRNP proteins preferentially associate with intron sequences and that this preference also helps the spliceosome distinguish introns from exons. However, as shown, at least some hnRNP proteins may bind to exon sequences but their role, if any, in exon definition has yet to be established. (Adapted from R. Reed, Curr. Opin. Cell Biol. 12:340–345, 2000.) 32 Domain structures of SR-family and SR-related proteins involved in pre-mRNA splicing. Large number of proteins that contain domains rich in alternating arginine and serine residues (RS domains).ASF/SF2 and SC35 are representative of a set of ten SR family proteins that contain one or two N-terminal RNA recognition motifs (RRM and a related CRRM motif) and a C-terminal domain that is rich in alternating serine and arginine residues (RS domain). SRm160 contains several domains of unknown function including the PWI motif (PWI), which is shared with other splicing-related proteins 31, polyserine-rich motifs (Ser), and serine, arginine and proline-rich motifs (SRP)21. 33 Models for exonic splicing enhancer (ESE) function. (a) The ‘U2AF-recruitment’ model for ESE function. SR proteins bound to an ESE are proposed to stabilize the binding of U2AF-65kDa to the polypyrimidine tract through an interaction mediated by U2AF-35kDa. SR proteins are also thought to form interactions across the intron by binding to the U1 snRNP-specific 70kDa protein (70K) and U2AF-35kDa. (b) A new model for ESE function in which U1 snRNP promotes two sets of cross-intron interactions: one of which is ESE-independent and the other is ESE-dependent. In contrast to the ‘U2AF-recruitment’ model, the ESE-independent interactions are sufficient for the stable binding of U2AF-65kDa to the polypyrimidine tract. This set of interactions probably involves BBP (the branchpoint- binding protein; also referred to as SF1 in mammals), which interacts with U2AF-65kDa, and the U1 snRNP-associated protein Prp40p. These interactions are highly conserved between yeast (S. cerevisiae) and mammals. The ESE-dependent interactions involve one or more SR proteins bound to an ESE, including SR-family and SRrelated proteins (e.g. hTra2), that bridge to one or more snRNP components through a splicing coactivator such as SRm160/300. Both sets of U1 snRNPdependent interactions are required for the stable binding of U2 snRNP to the branch site and the formation of a fully assembled spliceosome. Similar sets of interactions probably occur across exons, in accordance with the exon-definition model for splice-site selection. Different combinations of SR-family and SRrelated proteins that function in conjunction with different ESEs are likely to play a major role in determining regulated patterns of splice site selection in higher eukaryotes. 34 A Cascade of Regulated Pre-mRNA Splicing Controls Sexual Differentiation in Drosophila Activation by SR Proteins Rbp1 and Tra2 RequiresTra 35 Models of splicing silencing (a) Silencing by direct competition. A splicing-stimulatory factor, such as an SR protein, binds to an exonic splicing enhancer (ESE) (+; left). A splicing-inhibitory factor, such as a heterogeneous nuclear ribonucleoprotein (hnRNP), binds to an exonic splicing silencer (ESS) (; right). Because the ESE and ESS overlap, the binding of the positive and negative factors is mutually exclusive. If the positive factor has a higher binding affinity or higher concentration than the negative one, the alternative exon is included; if vice versa, then it is excluded. (b) Silencing by exon looping. A splicing inhibitory factor, such as an hnRNP protein, binds to duplicate intronic splicing silencer (ISS) elements present in the introns that flank an alternative exon. Dimerization of the bound protein brings the ISS elements into juxtaposition, causing the alternative exon to loop out and to be skipped by the splicing machinery. (c) Silencing by nucleation and cooperative binding. The alternative exon harbours ESE and ESS elements at some distance from each other. In the absence of an inhibitory factor, the ESS element remains unoccupied, and an SR protein can bind to the ESE and stimulate exon inclusion (+; left). An inhibitory factor (-; right) initially binds to a high-affinity binding site in the ESS, and nucleates cooperative binding of additional inhibitory molecules, which polymerize along the exon and displace the ESE-bound SR protein or prevent its initial binding, resulting in exon skipping. 36 Polypyrimidine tract-binding protein (PTB) is a nuclear factor that binds to the polypyrimidine tract of premRNA introns, where it is associated with negative regulation of RNA splicing and with exon silencing. Combinatorial control of splicing in the c-src N1 exon. Tissue-specific exons use a combination of positive and negative inputs to maintain their regulation. The N1 exon of the c-src gene is repressed by the PTB protein in non-neuronal cells where it binds to silencer elements on both sides of the exon and is thought to form an RNA looping complex. PTB binds in the downstream intron within a large complex, containing the KSRP and hnRNP F/H proteins. In neurons, the PTB is replaced with the related nPTB protein, and PTB binding to the upstream silencer elements is destabilized by an ATPdependent activity. Under these conditions, the downstream regulatory region can act as a splicing enhancer. The stimulatory proteins for this enhancer are not yet known, but require the UGCAUG element. The N1 exon itself also contains regulatory elements that bind to the ASF/SF2 and hnRNP A1 proteins. Similar models have been developed for most other tissue-specific exons that have been examined. 37 Quality Control of mRNA Schematic illustration of an “export-ready” mRNA molecule and its transport through the nuclear pore. As indicated, some proteins travel with the mRNA as it moves through the pore, whereas others remain in the nucleus. Once in the cytoplasm, the mRNA continues to shed previously bound proteins and acquire new ones; these substitutions affect the subsequent translation of the message. Because some are transported with the RNA, the proteins that become bound to an mRNA in the nucleus can influence its subsequent stability and translation in the cytosol. RNA export factors, shown in the nucleus, play an active role in transporting the mRNA to the cytosol . Some are deposited at exon-exon boundaries as splicing is completed, thus signifying those regions of the RNA that have been properly spliced. MESSENGER-RNA-BINDING PROTEINS AND THE MESSAGES THEY CARRY Gideon Dreyfuss*,V.Narry Kim‡ and Naoyuki Kataoka* NATURE REVIEWS | MOLECULAR CELL BIOLOGY VOLUME 3 | MARCH 2002 | p195 From sites of transcription in the nucleus to the outreaches of the cytoplasm, messenger RNAs are associated with RNA-binding proteins. These proteins influence pre-mRNA processing as well as the transport, localization, translation and stability of mRNAs. Recent discoveries have shown that one group of these proteins marks exon–exon junctions and has a role in mRNA export. These proteins communicate crucial information to the translation machinery for the surveillance of nonsense mutations and for mRNA localization and translation. 38 A Rôle de l’épissage dans la reconnaissance du codon stop prématuré. La règle de position stipule que lorsqu’un codon de terminaison est situé à plus de 50 nucléotides en amont de la dernière jonction exon-exon, alors il est reconnu comme prématuré (PTC) et l’ARNm est dégradé par NMD. Lorsque le codon de terminaison est situé à moins de 50 nucléotides de la dernière jonction ou dans le dernier exon (Ter), l’ARNm ne sera pas dégradé et il y aura arrêt de la traduction. (B) Un complexe protéique de 335kDa est déposé en amont des jonctions exon-exon lors de l’épissage du pré-ARNm. Ce EJC (‘ exon-exon junction complex’ ) favorise l’export de l’ARNm via l’interaction entre REF et TAP/p15, qui va à son tour interagir avec des composants du pore nucléaire (NPC). Dans le cytoplasme, seules les protéines Y14, Magoh, hUpf3 et RNPS1 restent associées à l’ARNm. Le EJC semble être le lien qui manquait entre l’épissage, l’exportation et la reconnaissance du PTC. 39 A model for the evolution of the EJC upon mRNA export. Pre-mRNA splicing deposits the EJC upstream of the splice junction. At this stage, the complex contains at least five proteins, SRm160, DEK, RNPS1, REF and Y14. After splicing, the TAP/p15 heterodimer and Upf3 join the complex in the nucleus. DEK probably dissociates (dashed arrow) before mRNA export to the cytoplasm, whereas SRm160 and the nucleocytoplasmic shuttling proteins RNPS1, REF, TAP/p15 and Upf3 are all likely to leave after mRNA export. Y14 remains stably associated with spliced mRNA in both compartments, while Upf2 only joins the complex in the cytoplasm. Question marks symbolize potential unidentified components. 40 Mécanismes de contrôle de qualité des ARNm. Après l’épissage, un résidu du complexe d’épissage (le complexe EJC) reste en place sur l’épissure. A. Si l’ARNm est «normal», le complexe EJC est chassé par le passage des ribosomes. Lorsqu’un codon stop «prématuré» précède l’épissure, les ribosomes restent en place et ne chassent pas le complexe EJC. Plusieurs cas de figures sont observés. B. le complexe EJC demeuré en place recrute un système de dégradation des ARNm 5’-3’ (non-sense mediated decay ou NMD). C. Les prémessagers «anormaux» s’agrègent près de leur site de transcription. D. Un épissage alternatif provoque la disparition du codon stop prématuré (non-sense mediated alternative splicing ou NAS). E. En l’absence de codon stop, le ribosome progresse jusqu’au bout de la queue polyA où il demeure arrêté et recrute le complexe de l’exosome qui dégrade l’ARNm de 3’-5’. 41 Model for the functional coupling of pre-mRNA splicing and nonsense-mediated decay. The exon–exon junction complex (EJC) assembles on messenger RNA as a result of splicing. TAP/p15 binds to components of the EJC and mediates the interaction of the mRNP with the nuclear-pore complex (NPC). During export, and immediately afterwards, the composition of the complex is changed as several of its components dissociate, leaving behind the remnants — the cytoplasmic EJC (cEJC), which includes Y14 and magoh, and possibly Upf3 and RNPS1 — at the same position as the complex was assembled in the nucleus. Upf2 probably binds to Upf3 after export. 42 Model for the role of the exon–exon junction on nonsense-mediated mRNA decay. In normal mRNAs, termination codons (Ter) are usually located downstream of the boundary; that is, 55 nucleotides upstream of the last exon–exon junction. So, translating ribosomes remove all the Y14–Upf2–Upf3 complexes (cEJCs) by the time of termination. During termination, Upf1 and the release factors (eRFs) are recruited and form the surveillance complex, which scans for the remaining Y14–Upf complex. Unless the surveillance complex encounters a Y14–Upf complex, translation proceeds to the next round. In the case of mRNA that contains a premature termination codon (PTC) more than 55 nt upstream of the last exon–exon junction, translation termination occurs before all the Y14–Upf complexes are removed. The remaining Y14–Upf complex interacts with the surveillance complex and triggers decapping of the mRNA. EJC, exon– exon junction complex; NMD, nonsense-mediated mRNA decay. 43 Types of RNA editing of mRNA creation of start codons by U insertion mRNA creation of new open reading frames by nucleotide insertion creation of stop codons by U insertion STOP AUG removal of stop codons by base conversions creation of start codons by C to U changes changes in encoded amino acids and splice site choice by base conversion creation of stop codons by C to U changes Substitution Editing Example: RNA editing of the human APO-B gene MEDECINE/SCIENCES 2002 ; 18 : 181-192 44







