view
advertisement

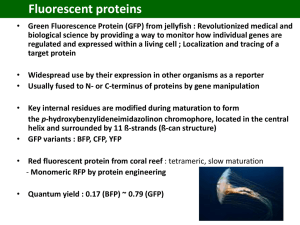
Chemistry of photoreceptors Contents • • • • • Introduction Red/Far red photoreceptors Blue light photoreceptors Blue/UV-A light photoreceptors Green Fluorescent protein(GFP) Introduction • Light is vital for a plant’s life - regulates plant growth and development. • Three major categories of photoresponses • Phototropism(Photosynthesis) • Photoperiodism(seed germination) • Photomorphogenesis(synthesis of chlorophyll) Ref: Phytochrome and photomorphogenesis, Harry Smith • What are photopigments ? Organic molecule which absorbs in the near UV-Visible range and upon absorption of the photon initiates a chemical reaction. Criteria • Absorption spectrum of the pigment should overlap with the wavelengths that are abundantly present in sunlight. • The pigment must have high extinction coefficient. • The excited state of the photopigment must have long lifetime. Ref: Light signals and the growth and development of plants - a gentle introduction, Pedro J. Aphalo Classification of photopigments Two types of photopigments Mass pigments • Absorbs large fraction of light. • Harvest energy (egs). Chlorophyll, flavonoids etc Sensor pigments • Absorbs small fraction of light. • Adjusts behaviour of plants. • red/far red, UV-A UV-B photoreceptors Ref: Aziz Sancar,Annu. Rev. Biochem., 2000(69), 31-67 Types of photoreceptors 1. Rhodopsins(visible) 2. Phytochromes(red/far red) 3. Xanthopsins(blue) 4. Cryptochromes(Blue/UV-A) “Primary photochemistry of photoreceptors activation is cis-trans isomerisation of the chromophore” Photoreceptors and their functions • Rhodopsins - Black & white and colour vision in higher animals • Light harvesting complex(LHC) - Photosynthesis in plants. • Phytochrome - Photomorphogenesis in plants • Cryptochrome - Regulation of circadian rhythm in living organism. • Photolyase - Repair of DNA damage in drosophila, xenopus etc. Classification of photoreceptors CHROMOPHORES Classes PHOTOSENSORS PHOTOCHEMISTRY FAMILY example structural unit R1 N Tetrapyrroles phytochromobilin R2 Phytochromes trans Cis Rhodopsins trans Cis Xanthopsins trans Cis N O Polyenes retinal Coumaric acid 13 11 12 14 N H O -O S- Aromatics R Flavin N N O Cryptochromes electron transfer N N O Ref: Klass J. Hellingwerf, J. Photchem. Photobiol. B, 2000(54), 94-102 Phytochrome Red/far red photoreceptor Introduction • Phytochrome was discovered by Borthwick and Hendrick while trying to understand the photoperiodic control of flower formation and light regulation of seed germination. • Phytochrome is the best known light receptor in all higher plants, algae, mosses etc. • Phytochrome absorbs visible light both in blue and red regions. • Energy of the absorbed light is then converted to signals which exert photomorphogenic controls (i.e., control of plant growth). • Phytochromes are responsible for controlling seed germination, stem elongation, leaf expansion, synthesis of chlorophyll etc. Structure of phytochrome • Phytochrome is a protein with molecular weight of 124 kDa • Phytochrome molecule has a tetrapyrrole chromophore absorbing in the red region. • The chromophore is bound to sulfur of cystein-321. • The structure of the chromphore(tetrapyrrole) was confirmed by oxidative degradation by chromic acid. • Phytochrome(P) can exist in two relatively stable forms 1. Physiologically inactive form(Pr)-red absorbing form 2. Physiologically active form(Pfr)-far red absorbing form Photochemistry of Phytochrome chromopho 660 nm Pfr Pr Physiological reaction 730 nm Ref: W. Rudiger, F. Thummler., Angew. Chem. Intl. Ed., 1991(30), 1216-1228 Mechanism for the conversion of Pr to Pfr • Z, E isomerisation of the chromophore C16 C5 C15 C10 red light far red light • Pr chromophore - 5Z, 10Z, 15Z Pfr chromophore - 5Z, 10Z, 15E Evolution of Phytochrome function • Phytochrome has five genes (phyA, phyB, phyC, phyD, phyE) • Phytochrome regulates almost all phases of plant development. “Single input/multiple ouput” sensory systems How does phytochrome work ? • Sunlight(660 nm) converts Pr to Pfr, the Pfr moves from the cytoplasm into the nucleus. • It binds then to a protein called PIF3(“phytochrome-interacting factor 3”) which is a helix-loop-helix protein. • The complex of two then binds to the promoters containing the sequence CACGTG, GTGCAC. • These promoters are found in genes that themselves encode other transcription factors and initiate transcription. • Exposure to far red light converts the Pfr back to Pr form and PIF3 dissociates from Pfr. Photoactive Yellow Protein(PYP) Blue light photoreceptor Introduction • PYP was discovered almost 20 years ago in Ectothioshodospira Halophila. • PYP belongs to Xanthopsins family - a blue light photoreceptor protein. • PYP is the biopigment responsible for photophotactic activity of the purple phototropic bacteria. • Purple phototropic bacteria(H. halophilia) is not immune to UV light. It is attracted by infra red light(for photosynthesis) but moves away from blue light. Photoresponses of R. Centenum. Positive movement towards infrared light Negative movement towards visible light. Accumulation pattern of cells of E.halophilia in response to blue light illumination. Structure of Photoactive yellow protein • 125 amino acid residues with molecular weight of ~ 14 KDa -O O S Protein Chromophore structure p-hydroxy thiocinnamate Structure of PYP from Halorhodospira halophila Ref: K. J. Hellingwerf, J. Hendriks, T. Gensch, J. Phys. Chem. A, 2003(107), 1082-1094 • The chromophore is linked through a thiol ester linkage to Cys69. • Negative charge is stabilized via a hydrogen bonding network with residues Try42, Glu46, Thr50. • UV/Vis absorption(solid line) and fluorescence emission spectrum λmax = 446nm φfluo = 0.002 The PYP Photocycle • PYP* - excited state species • pR - redshifted photoproduct • pB - blueshifted photoproduct • More detailed analysis of low temperature spectroscopic experiments revealed branched pathways. A Comprehensive PYP photocycle • Intermediates of photocycle of PYP were determined at - 12°C. • At very low temperatures(-120 º C) another intermediate PYPBL was found. • Analysis of crystal structures of photoproducts indicated that the key feature is the flipping of the carbonyl group of the thioester linkage. • Signalling state is pB. • pB interacts with transducer protein. Cryptochromes Blue light/UV A radiation photoreceptors Introduction • Cryptochromes are receptors of blue light and UV A radiation. • Blue light responses are present in all species tested from bacteria to plants and animals. • Various responses observed are photoactivation in bacteria, phototropism, photomorphogenesis in plants, circadian rhythm in fungi and drosophila. Types of Cryptochromes 1. Photolyase - involved in DNA repair mechanism Drosophila, virus. 2. HY, phototropin - regulates phototropism in plants 3. CRY1, CRY2 - mediate circadian clock in humans. Biological clocks • Most of our activities follows a set of pattern - “Biological rhythm”. • Biological rhythm is an expression of internal clock(biological clock). • Circadian rhythm - biological functions with periodicity of approximately 24 hours. • Circadian rhythms are observed in most living organisms ranging from cyanobacteria to humans to plants. • hCRY1 and hCRY2 are the proteins which mediate the biological clock. • hCRY1 is 586 amino acid long 66 kDa protein. hCRY2 is 593 amino acid long 67 kDa protein. Photolyase • Photolyase is a 55 to 64 KDa protein. • Repairs UV induced DNA damage in a reaction dependent on near UV to blue light(350-450 nm). Ref: H. Komori, R. Masui, S. Kuramitsu, S. Yokoyama, T. Shibata, Y. Inoue, K. Miki, PNAS, 2001(98), 13560-13565 DNA damage by UV light • Absorption of UV light by DNA results in the formation of triplet state of thymine. • Thymine can return to ground state or can undergo a pericyclic reaction(2+2 cycloaddition) with adjacent pyrimidine ring of thymine or cytosine. O O NH O OR N O UV O O O P NH N H O O O O HN NH O N O OR OR O- photolyase N O O O near UV-visible H O P O- OR O Photoreactivation • Damaged DNA can be repaired by photolyases activated by blue/UV A light. • Chromophore in photolyase responsible for DNA repair. a) Flavin adenine dinucleotide(FAD) in their anionic form b) Methenyl tetrahydrofolate(MTHF) or 8-Hydroxy 5 deaza riboflavin. Electronic energy transfer • The two chromophores folate(MTHF) and flavin(FADH-) have different characteristics. Folate(MTHF): ε = 25000 M-1cm-1 Flavin(FADH-): ε = 5000 M-1cm-1 • Dependence of light penetration into the cells shows that folate excels flavin as a light absorber. • Energy transfer from folate(donor) to acceptor(flavin) occurs by Forster type energy transfer. kET ∝ k2J/R6 k2 = orientation of interacting dipoles J = spectral overlap R= distance between the dipoles Mechanism • The photoreactivation enzyme(photolyase) binds to pyrimidine dimers in the DNA(Kasso = 2.6 x 108 M-1). • Upon exposure to light, MTHF absorbs a photon(photoantenna) and transfers excitation energy to flavin. • Flavin transfers an e- to the DNA photoproduct: the cyclobutane ring of the pyrimidine is broken to generate two pyrimidine rings. • Back electron transfer restores the FADH- and enzyme dissociates from DNA. • Quantum yield of the reaction is found to be 0.5-0.9. Ref: J. E. Hearst, Science, 1995(268), 1858-1859 Green Flurorescent Protein • GFP was first found in a jellyfish named Aequorea which fluoresced green when irradiated with UV light. QuickTime™ and a GIF decompressor are needed to see this picture. Chromophore structure p-hydroxy benzylidene • Chromophore is buried under compact, rigid protective structure - highly stable and has high quantum yield. • Isolated chromophore is not fluorescent whereas the folded protein is fluorescent in nature. • Chromophore formation - Autocatalytic postransational cyclisation. • Depending on the chromophore, GFP can be classified into seven classes. Mechanism of chromophore formation • Chromophore formation is the bottleneck of protein folding. Mechanism of fluorescence • GFP has two absorption peaks - at 398 nm and 475 nm. • Emission at 503 to 509 nm when excited at 398 nm and emits at 482 nm when excited at 475 nm. • Two absorption states: Chromophore in phenolic neutral form(A state) Chromophore in phenolate anionic form(B state) • Excited state proton transfer from chromophore to Glu222. Applications of GFP • The applications of GFP are as Fusion tags, Reporter gene, biosensors, photobleaching, FRET etc. 1. Fusion Tag • Fusion between cloned gene and GFP can be used to visualize dynamic cellular events. 2. Reporter Gene • Gene expression can be monitored by using a GFP gene that is under the control of the promoter of interest. • The change in the intensity of fluorescence of GFP directly indicates the level of gene expression. 3. Fluorescence Resonance Energy Transfer • Folding and Unfolding dynamics of protein can be studied using GFP. CFP, YFP is connected to metal lathionein(Zn) Add EDTA Unfolding - Zn binds with EDTA (F535nm/F480nm) decreases 4. Biosensors and Bioelectronics • Genetically modified plants are being used as biosensors. • Biosensors responsive to copper and TNT has been designed. • Concept of photobleaching is now been exploited for bioelectronics and memory storage devices. 5. How does a non-scientist looks at GFP ? GFP-bunny GFP-monkey Conclusion • Photochemistry plays a important role in everyday life starting from control of plant growth, photosynthesis in plants, black and white and colour vision in higher animals.