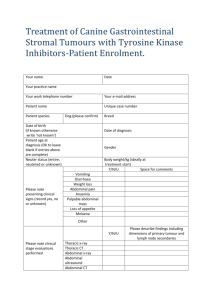
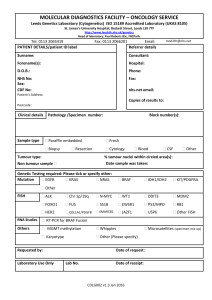
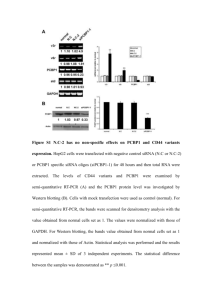
Supplementary Information (doc 68K)
advertisement

Supplementary Table 1: Clinico-pathological data of primary CRCs Parameter Category n (%) Age (years) Median Range Male Female Alive Dead (cancer related) Dead (unrelated causes) Unknown Adenocarcinoma Mucinous adenocarcinoma Columnar adenocarcinoma Signet ring mucinous adenocarcinoma Unknown Well differentiated Moderately differentiated Poorly differentiated Unknown Colon Rectal Unknown A B C1 C2 D Unknown 0 (Tis) 1 2 3 4 Unknown Negative Positive Unknown 72 57-89 257 (57%) 192 (43%) 167 (37%) 221 (49%) 60 (13%) 1 382 (85%) 49 (11%) 4 (1%) 6 (1%) Sex Status Histological type Histological grade Tumour site Duke’s stage TNM stage Extramural vascular invasion 8 (2%) 28 (6%) 345 (77%) 67 (15%) 9 (2%) 230 (52%) 177 (39%) 42 (9%) 66 (15%) 175 (39%) 133 (30%) 20 (4%) 52 (11%) 3 (1%) 3 (1%) 67 (15%) 172 (38%) 149 (33%) 51 (11%) 7 (2%) 219 (49%) 121 (27%) 109 (24%) The patients had a median age of 72 years (range 45–80 years) with a median follow-up of 37 months (range 0-116). Fifty percent of patients were male while 43% were female. Regarding the histological type, the majority of tumours 345 (77%) were moderately differentiated adenocarcinomas, 28 (6%) well differentiated tumours, and 67 (15%) were poorly differentiated. Vascular invasion was present in 121 (27%) of cases, while 219 (49%) had no evidence of vascular invasion, and this information was not available for the remaining 109 (24%) of cases. A total of 221 (49%) patients died from CRC, 60 (13%) died due to other causes, while 167 (37%) of patients were alive at the end of follow-up. 1 Supplementary Table 2: Association of nuclear Cten expression with tumour metastasis Type of tumour deposit Low nuclear Cten expression High nuclear Cten expression Primary Metastasis 36 25 4 15 A series of 40 paired primary and metastatic tumours were evaluated for Cten expression by immunohistochemistry. Positive nuclear expression of Cten was associated with liver metastasis (Fisher’s exact test, p=0.002) 2 Materials and methods Tissue microarray analysis (TMA) and Immunohistochemistry (IHC) A tissue microarray analysis (TMA) of primary tumours was prepared from a series of 462 cases of primary operable CRC from patients who underwent elective surgery between 1st January 1993 and 31st December 2000 at the Nottingham University Hospitals, Queen’s Medical Centre, Nottingham as has been previously described(1, 2). This resource has been well characterised and a prospectively maintained database was used to record relevant clinical and pathological data. The length of follow-up was determined from the date of primary tumour resection, with surviving cases censored for analysis on the 31st December 2003. Disease-specific survival (DSS) was used as the primary end-point. A TMA was also prepared from a series of 40 patients from whom tumour material from both primary tumour and corresponding hepatic metastasis was available. Immunohistochemistry (IHC) for Cten was performed with full local ethical approval. Slides were stained using the water bath antigen retrieval method with EDTA Buffer (Sigma) as previously described(3). The primary antibody Cten (Abcam,ab57940.1:75), was applied for 45 minutes at room temperature and diaminobenzidine (DAB) (Sigma) was used as the chromogen. Slides were then counterstained with haematoxylin (Dako). Evaluation of IHC The IHC was scored by two pathologists (AA and WF) using the Hscoring system. Briefly, the slides were assessed for the intensity of tumour cell staining ranging from zero to three, (zero = no staining, one = weak, 2 = moderate, three = strong) and for the percentage of tumour cells staining within each intensity category. The two numbers obtained were multiplied and their product represents the H-score. Cell culture and transfection The CRC cell lines used in this study were kindly donated by Prof I Tomlinson. All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% foetal calf serum (Invitrogen) and penicillin/streptomycin (respectively at concentrations of 100 units per ml 3 and 100g per ml, Invitrogen) in 5% CO2 in a humidified atmosphere. In order to induce Cten expression, cells were transfected with a CMV promoter driven expression construct containing Green Fluorescent Protein (GFP) tagged Cten (GFP-Cten) using Lipofectamine 2000 (Invitrogen) as previously described(3). Control cells were transfected with a GFP empty vector. In order to knock down Cten and ILK, cells were transfected with small interfering RNA (siRNA) duplexes using Lipofectamine 2000 (Invitrogen) as previously described(4). The cells were transfected with each siRNA duplex at a final concentration of 100nM and compared with cells transfected with sequence scrambled control (ssc) duplexes (i.e. duplexes with the same base composition arranged in a random order). Sequences for all the duplexes used are available from the authors. In order to functionally evaluate Cten, a number of different cellular conditions were created using a series of co-transfections (see results). For cotransfection of GFP-Cten with an siRNA, the protocol for siRNA transfection was used with appropriate amounts of plasmid mixed in with the siRNA prior to adding to Lipofectamine 2000. For simultaneous knockdown of two separate targets, each of the relevant siRNAs were admixed to maintain a final concentration of 100nM for each duplex. Cells were analysed 72 hours later by Western blot or functional assays. Western blotting Whole cell extracts were prepared using lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1% TritonX-100, 0.5% sodium deoxycholate, 1 mM EDTA, 0.1% SDS, supplemented with protease and phosphatase inhibitors (Sigma)). 30g protein was loaded on a 10% SDS–PAGE gel and transferred onto PVDF membranes by semi-dry transfer. After blocking, membranes were incubated overnight at room temperature with the indicated primary antibody. Antibodies were used at the following dilution: anti-Cten (Sigma, WH0084951M1,1:1000), anti-ILK antibody (Abcam, ab19025, 1:500) and mouse anti--actin (Sigma, 1:2000). After three washes in TBS/Tween-20 (0.05%), blots were incubated for 1 hr at room temperature with the appropriate horseradish peroxidaselinked secondary antibody. After three further washes, detection was 4 performed using the Enhanced Chemiluminescence Kit (Pierce). Bands were visualised using X-Ray films (Kodak). Cell Migration/ Invasion Assays Transwell cell migration was measured using a Boyden chamber containing a polycarbonate filter with an 8m pore size (Costar). 5 X 104 cells were seeded and cell migration was assessed after 48 hr by fixing the cells attached to the lower surface in 10% methanol for 30 min, staining for 30 min with methylene blue and manually counting the stained cells. Cell invasion was measured in the same way as described for migration, except that prior to cell seeding the upper chamber was prepared by coating the filter with 100l Matrigel (5 mg/ml; BD Biosciences) and the cells attached to the lower surface were fixed, stained and counted. All assays were performed at least twice in triplicate. In vivo metastasis model Male MF1 nude mice (Harlan-Olac) were used. Animal procedures were run under the Home Office project PPL 40/2962 following local ethical approval. For analysis of tumor growth and metastasis, the CRC cell line HCT116 was stably transfected with either a GFP-Cten expression vector (HCT116GFP-Cten) or empty vector control (HCT116evc) expressing GFP only. Cells were injected into the spleens of the animals using standard techniques. Briefly, a left abdominal flank incision was made and 1 x106 cells were injected into the subcapsular region of the spleen of nude mice (n=10 in each group). On day 42, mice were sacrificed by approved S1 method and the spleen, liver and lung were dissected and examined for orthotopic tumors and distant metastases. Organs were formalin-fixed and paraffin-embedded and 4m sections were stained with Haematoxylin and Eosin. All tissue sections were scanned at x 20 using the NanoZoomer 2.0 (Hamamatsu Corporation, Japan) and the tumour volume was obtained using the NanoZoomer Digital Pathology Image (ndpi) software (Hamamatsu Corporation, Japan). Statistical analysis 5 Data were analysed using SPSS 16.0 statistical software (SPSS Inc. USA). For the IHC, determination of the optimal Cten cut-off was performed using Xtile bioinformatics software (version 3.6.1, 2003–2005, Yale University, USA) (5). This program basically divides the total patient cohort randomly into two separate equal training and validation sets ranked by patients’ follow-up time. The optimal cut-points are determined by locating the brightest pixel on the Xtile plot diagram of the training set and the scoring are dichotomised into “low Cten” and “high Cten”. Associations between Cten expression (i.e. low or high) and clinicopathological parameters were evaluated using Pearson 2 test. Patients whose deaths were attributed to colorectal cancer, were considered in the disease-specific survival calculations. Deaths as a result of non-colorectal cancer related causes without evidence of recurrence were censored at the time of death. Survival curves were analysed by the Kaplan– Meier plot with a log rank test to assess significance. Multivariate Cox proportional hazards model was used to test the statistical independence and adjust for confounders. A two-tailed p-value of <0.05 was considered significant. All other evaluations were done using the unpaired two-tailed Student’s t–test and p< 0.05 was considered to indicate a significant difference. 1. Ullenhag GJ, Mukheriee A, Watson NFS, Ai-Attar AH, Scholefield JH, Durrant LG. Overexpression of FLIPL is an independent marker of poor prognosis in colorectal cancer patients. Clinical Cancer Research. 2007;13(17):5070-5. 2. Ahmed MA, Al-Attar A, Kim J, Watson NF, Scholefield JH, Durrant LG, et al. CD24 shows early upregulation and nuclear expression but is not a prognostic marker in colorectal cancer. J Clin Pathol. 2009 Dec;62(12):111722. 3. Albasri A, Seth R, Jackson D, Benhasouna A, Crook S, Nateri AS, et al. C-terminal Tensin-like (CTEN) is an oncogene which alters cell motility possibly through repression of E-cadherin in colorectal cancer. J Pathol. 2009 May;218(1):57-65. 4. Elsaba TM, Martinez-Pomares L, Robins AR, Crook S, Seth R, Jackson D, et al. The stem cell marker CD133 associates with enhanced colony formation and cell motility in colorectal cancer. PLoS One.5(5):e10714. 5. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clinical Cancer Research. 2004;10(21):7252-9. 6