Restriction Digests
advertisement
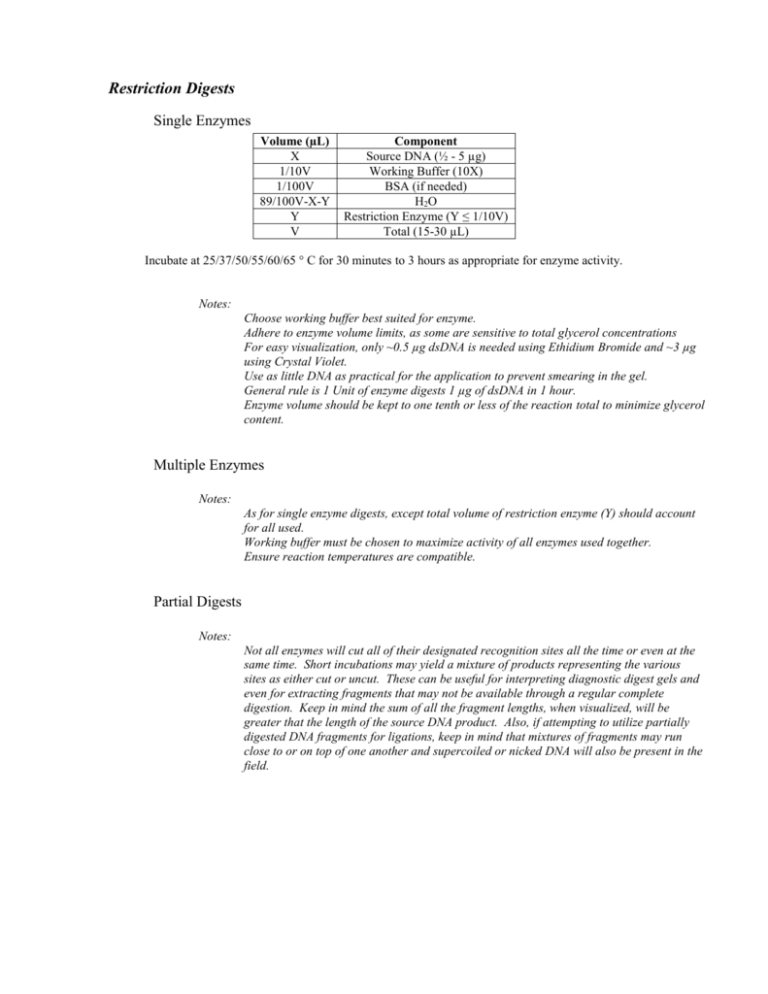
Restriction Digests Single Enzymes Volume (µL) X 1/10V 1/100V 89/100V-X-Y Y V Component Source DNA (½ - 5 µg) Working Buffer (10X) BSA (if needed) H2O Restriction Enzyme (Y ≤ 1/10V) Total (15-30 µL) Incubate at 25/37/50/55/60/65 ° C for 30 minutes to 3 hours as appropriate for enzyme activity. Notes: Choose working buffer best suited for enzyme. Adhere to enzyme volume limits, as some are sensitive to total glycerol concentrations For easy visualization, only ~0.5 µg dsDNA is needed using Ethidium Bromide and ~3 µg using Crystal Violet. Use as little DNA as practical for the application to prevent smearing in the gel. General rule is 1 Unit of enzyme digests 1 µg of dsDNA in 1 hour. Enzyme volume should be kept to one tenth or less of the reaction total to minimize glycerol content. Multiple Enzymes Notes: As for single enzyme digests, except total volume of restriction enzyme (Y) should account for all used. Working buffer must be chosen to maximize activity of all enzymes used together. Ensure reaction temperatures are compatible. Partial Digests Notes: Not all enzymes will cut all of their designated recognition sites all the time or even at the same time. Short incubations may yield a mixture of products representing the various sites as either cut or uncut. These can be useful for interpreting diagnostic digest gels and even for extracting fragments that may not be available through a regular complete digestion. Keep in mind the sum of all the fragment lengths, when visualized, will be greater that the length of the source DNA product. Also, if attempting to utilize partially digested DNA fragments for ligations, keep in mind that mixtures of fragments may run close to or on top of one another and supercoiled or nicked DNA will also be present in the field.