nph12091-sup-0001-FigS1-S6-TableS1-S2
advertisement

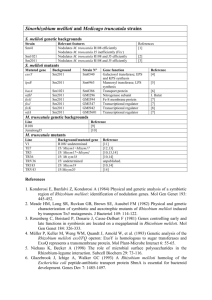
Supporting Information Figs S1–S6 and Tables S1 & S2 Table S1 Primers used in this study. Primers used for MTR_4g085800 mutant screening (a), splicing variant cDNA synthesis (b) qRT-PCR (c), and the pMB132 construct (d). Table S2 dnf2 nodule cells have a reduced endoreduplication index. Plant line, nodule age, DNA content and endoreduplication index are indicated for roots (r) and for nodules (n). Results are given for dnf2-2 and dnf2-4. Similar results were obtained in independent experiment using dnf2-3 line. Fig. S1 Medicago truncatula DNF2 is a conserved protein containing a phosphatidylinositolspecific phospholipase C X domain. The deduced amino acid sequence of Medicago truncatula DNF2 was aligned with orthologous proteins from Lotus japonicus and Arabidopsis thaliana using ClustalW2 (www.clustal.org/). Predicted signal peptides (http://bmbpcu36.leeds.ac.uk/prot_analysis/Signal.html) determined are with represented in SIG-PRED bold. The Phospholipase C, phosphatidylinositol-specific, X domain (IPR000909) defined by Prosite (http://prosite.expasy.org/) is underlined. An * (asterisk) indicates positions which have a single, fully conserved residue. A : (colon) indicates conservation between groups of strongly similar properties. A . (period) indicates conservation between groups of weakly similar properties. Fig. S2 Medicago truncatula dnf2-2, dnf2-3 and dnf2-4 mutants are strongly altered in DNF2 expression. 1 DNF2 expression was investigated in dnf2-2, dnf2-3 and dnf2-4 mutant nodules by qRT-PCR. Primers dnf2c and 217Ase4-2 were used to amplify DNF2 cDNA prepared from 27 dpi nodules. DNF2 mRNA was undetectable in dnf2-2 and dnf2-4, demonstrating that these mutants are knock-out mutants. dnf2-3 shows a strongly reduced signal, hard to differentiate from the negative control. Error bars represent standard errors. Fig. S3 dnf2 is required to support nifH expression and nitrogen fixation. (a) Medicago truncatula R108 wild-type and dnf2-4 plants were grown in symbiotic conditions inoculated with Sinorhizobium meliloti strain Sm1021. dnf2-4 plants are smaller than wild-type and display chlorosis and accumulation of anthocyanins. Scale bar: 2.5 cm. (b) and (c): M. truncatula wild type R108 (b) and dnf2-4 (c) mutant nodules induced with a S. meliloti Rm41 derivative strain harboring a nifH::lacZ fusion. 15 days post inoculation (dpi) nodule slices were stained to reveal beta-galactosidase activity, and thus, nifH promoter activity. Blue pigmentation was observed only in R108 wild type, illustrating the incapacity of the dnf2-4 mutant to support nifH transcription. Scale bars: 200 µm (d) Acetylene reduction assays were conducted on entire plants using R108 wild-type and dnf2-4 mutant 27 dpi. Acetylene reduction activity per plant expressed as arbitrary units (A.U.) was only detectable in wild-type plants. Error bar represents standard error. Fig. S4 Occurrence of brownish nodules in dnf2 nodules elicited by Sinorhizobium meliloti Rm41 or Sm1021 strains. Number of total nodules formed by dnf2-4 plants is higher than those formed by wild-type plants. Wild-type and dnf2-4 nodules were counted on five pools of nine plants each. White bars represent non-fixing nodules. Pink bars represent N2-fixing nodules. Brown bars 2 represent brownish nodules. Note that brownish nodules are present only on the mutant line. For convenient comparison data of Rm41 inoculated plants are reproduced from Figure 2. Fig. S5 DNF2 is alternatively spliced in legume plants. (a) The DNF2 gene contains eight exons represented by black boxes. The third exon is depicted as a black and grey box. The grey part in the 3’ part of the third exon contains four alternative splicing sites. The first intron is represented by a white box and all other introns are represented by black lines. The most abundant transcript, corresponding to TC119183, represents 67% of total transcripts isolated by cloning full length cDNAs and encodes a 334 amino acids peptide. The sequence retained at the 3’ end of the third exon in this form is indicated in (1). The longest transcript retains the first intron (white box) and encodes a 360 amino acids peptide. It represents 16% of total transcripts and the sequence retained at the 3’ end of the third exon is also indicated in (1). The splicing form (3) generates an in frame transcript representing 10% of total transcripts and encodes a 325 amino acids peptide. The two other splicing form indicated in (2) and (4) were found in the remaining 6% of the total transcripts. They putatively encode 310 amino acids and 303 amino acids peptides using an alternative ATG codon represented in grey. A rare splicing form lacking the third exon and generating a 296 amino acids peptide with a signal peptide was also found (not represented). (b) Comparison of the splicing sites in the 3’ region of the third exon in Medicago, Phaseolus and Lotus DNF2. The exon sequences are highlighted in grey and the intron splicing sites in green. In Medicago the 1st and the 3rd splicing sites are not in frame and generate transcripts encoding 303 and 310 amino acids peptides respectively. The 2nd and 4th splicing sites are in frame and generate transcripts encoding 325 and 334 amino acids peptides respectively. The splicing sites in the Phaseolus gene were determined using the P. vulgaris ESTs CV537130, CV534814, CV534298, CV536519, CV535147, CV533809 and CV535433. Some of these 3 ESTs may represent partially spliced mRNA (pre-mRNA) because they retain some intronic sequences. The Lotus sequence is given for comparison but putative splicing sites are not experimentally supported. Note the sequence conservation in this region for the three species. Fig. S6 Impairment of bacteroid differentiation in dnf2 nodules. For these experiments the Sm1021 strain of Sinorhizobium meliloti was used. A culture of Sm1021 was used as the free-living rhizobia sample. Bacteroids were extracted from wildtype and dnf2-4 nodules inoculated with Sm1021. (a to c) DAPI staining of free-living rhizobia (a), wild-type bacteroids (b), and dnf2-4 bacteroids(c). Scale bars: 25 µm. (d to f) Cytometric analyses of free-living rhizobia (d), wild-type bacteroids (e) or dnf2-4 bacteroids (f). Bacterial populations of the different samples are represented on scatter plot. For the cytometric analyses DNA contents measured by DAPI staining are indicated on the Y-axis and granularity of bacteria is indicated on the X-axis (granularity reflects the cellular structure of the bacteria). The population density is indicated by a color range scale below the graph. dnf2-4 bacteroids appear as intermediate between free-living rhizobia and wild-type bacteroids. 4 Primer name MTR_4g085800 Starta MTR_4g085800 Stopa LTR4a LTR6a MERE-1a MERE-4a 4-1b 4-3b 2-bb 2-db ex5-6b AUGb TC94217-Fc TC94217-Rc TC100436-Fc TC100436-Rc MtRBP1-Fc MtRBP1-Rc 217Ase4-2c dnf2cc Fusprom1d Fusprom2d Sequence AGCTAATGGAGGTAAGTGTGATT CTGAACTCACACAAACAAATTTCGA TACCGTATCTCGGTGCTACA GCTACCAACCAAACCAAGTCAA AGCCCTTTTGTCAAACATGTATTA CCTCTTTTCTCGTCTCTTATTT AGCTAATGGAGGTAAGTGTGATT CTGAACTCACACAAACAAATTTCGA TGCCTGGAATGCAACTCACAAA GGATGTCTTCAATCGTAGATAATGA TTAATGGCAGGTTGAAAGGCGGTA ATAGCTAATGGAGAATTACCTTCTAA CATTGTTGGAGGTGTTGGCCTT AGTAACCCTCTATAGCCTTGAA TGTCCCTGCCAACAATGAGCA CAACCAATACTTGGTTCCATCAT AGGGGCAAGTTCCTTCATTT AAACGGACGGAAAATGTGAG CGACACCGAACTGAGATAGTCA AGGCAATGCGTTCAGAAGCCT CTGCCCAGCTGTTGG CGCGCCAACAGCTGGGCAGCATG Table S1 5 Medicago truncatula Sinorhizobium meliloti WT R108r dnf2-4r WT R108n dnf2-4n WT R108n dnf2-2n Sm1021 Sm1021 Sm1021 Sm1021 Sm1021 pXLGD4 Sm1021 pXLGD4 age (dpi) 28 28 28 28 17 17 2C (%) 48,1 47,6 16,3 16,5 10,6 22 4C (%) 50,4 50,7 65,3 69,2 67,1 59,5 DNA content 8C (%) 16C (%) 1,5 0 1,7 0 7,4 5,2 9,9 2,1 9,1 5,8 11,3 3,8 32C (%) 0 0 5,7 2,1 6,59 3,1 64C (%) 0 0 0,3 0,2 0,9 0,4 E.I. 0 0 39,9 15,7 48,26 25,8 Table S2 6 L.japonicus BAD12493 M.truncatula DNF2 A.thaliana NP_564553.1 ----MENYLLIATLVSASLVFGCYYILVQIAETCSRDINDCDLGLQCLECHS----QNRC 52 ----MENYLLIATLISASLVFGCY-VLVQIGETCSRDVNDCGTGLQCLECNS----QSRC 51 MFQRLLLFLLIALLLQSSFLLEIS-SALKEGKTCITNSN-CDAGLHCETCIANTDFRPRC 58 : :**** *:.:*::: :: .:** : * *. **:* * : : ** L.japonicus BAD12493 M.truncatula DNF2 A.thaliana NP_564553.1 TRIRTISPTSKVMELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGV 112 TRVRTSSPISKVMELPFNHYSWLTTHNSYASRAANLSIDSKISSVMNQEDSITDQLRNGV 111 SRTQPINPITKAKGLPFNKYSWLTTHNSFARLGEVSRTGSAILAPTNQQDSITSQLNNGV 118 :* :. .* :*. ****.*********:* . .* : : **:****.**.*** L.japonicus BAD12493 M.truncatula DNF2 A.thaliana NP_564553.1 RGLMLDMWDYEDTIWLCR---GPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHV 169 RGIMLDMHDYYGDIWLCR---GPCTIFTAFQPAINVLREINTFLTRHRTEIVTVFIKDRV 168 RGFMLDMYDFQNDIWLCHSFDGTCFNFTAFQPAINILREFQVFLEKNKEEVVTIIIEDYV 178 **:**** *: . ****: *.* :*:****:*:*:*...** : *::*::*.* * L.japonicus BAD12493 M.truncatula DNF2 A.thaliana NP_564553.1 TSGNGVNKVFDKARLRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGI 229 TSPNGVNKVFNKAGLRKFWFPVYKMPKNGSDWLTVKKMLRMNHRLIVFTSNATKEASERI 228 KSPKGLTKVFDAAGLRKFMFPVSRMPKNGGDWPRLDDMVRKNQRLLVFTSDSHKEATEGI 238 .* :*:.***: * **** *** :*****.** :. *:* * **:****:: :**:* * L.japonicus BAD12493 M.truncatula DNF2 A.thaliana NP_564553.1 AYEWNYVVESQFGNVGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSS 289 AYEWNYVVENKYGNDGMGRGHCLHRAESYPMNTTTKSLVLMNYYRNVLN-SNEACRDNSS 287 AYQWKYMVENQYGNGGLKVGVCPNRAQSAPMSDKSKSLVLVNHFPDAAD-VIVACKQNSA 297 **:*:*:**.::** *: * * :*.:* **. :*****:*:: :. : **::**: L.japonicus BAD12493 M.truncatula DNF2 A.thaliana NP_564553.1 PLIAMMHVCFRAAGNRWPNFIAVDFYKRGDGGGAPEALDLANRNLIRL------------ 337 PLIRKMHTCYKDAGNRWPNYIAVDFYKRGDGGGAPEALDVANRNLFV------------- 334 SLLESIKTCYQAAGQRWPNFIAVDFYKRSDGGGAPQAVDVANGNLICGCDNFAACKADGK 357 .*: ::.*:: **:****:********.******:*:*:** **: L.japonicus BAD12493 M.truncatula DNF2 A.thaliana NP_564553.1 --CG 359 Fig. S1 7 Fig. S2 8 Fig. S3 9 Fig. S4 10 Fig. S5 11 Fig. S6 12