Characterization of Wolbachia pipientis Infections in Arthropods
advertisement

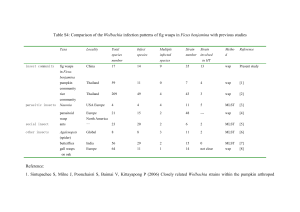
Characterization of Wolbachia pipientis Infections in Arthropods Native to Various Coastal New Jersey Habitats Joshua D. Nelson, Felicity Bennett, Lee Kerkhoff, PhD Submitted to Marine Biological Labs to fulfill the requirements for the MBL Summer Envisionship Program Introduction Wolbachia are an abundant and highly diverse group of endosymbiotic alphaProteobacteria, related to the Anaplasma, Erlichia, and Rickettsia genera, that was first described as a parasite in the mosquito Culex pipientis by Hertiz and Wolbach in 1924. After its discovery, Wolbachia went virtually unstudied until the early 1970s (1-3, 8, 9). Wolbachia was considered somewhat rare until the 1990s, when molecular biological techniques allowed investigators to discover that Wolbachia were widespread and very abundant in diverse hosts including arthropods and nematode worms. Since then, Wolbachia bacteria have been studied extensively, aided largely by further advances in molecular biology methods(1, 2, 5, 13, 18, 25). To date, eight distinct Wolbachia “supergroups” have been characterized, designated by letters A through H, based on 16S rRNA as well as functional gene sequence data (1-7). Wolbachia has become known as the most widespread obligatory endosymbiont on the planet, and field sampling and statistical analyses estimate that 20 to 60% of arthropod species are infected (10, 25). Despite recent advances in knowledge about Wolbachia, relatively little is known about the phylogenetic and evolutionary history of these bacteria, due to lack of DNA sequence data and challenges in identifying, or “strain-typing” these bacteria (2, 5). Baldo et al (2006) recently described multi-gene approach identifying Wolbachia strains using five genes that are highly conserved and that is ubiquitous to the Wolbachia genus, as well as a gene marker for a Wolbachia surface protein (2). This work has led to the formation of a Wolbachia strain-typing database that continues to grow. These amazing and diverse endosymbiotic bacteria are transmitted vertically through host germ cells and have evolved endosymbioses ranging from mutualism to reproductive parasitism. For example, Wolbachia have evolved mutualistic relationships with filarial nematode hosts, which depend on Wolbachia for survival and likely protect nematode worms from host immune systems (5, 18, 23, 25). In contrast, Wolbachia have evolved reproductive parasitism in arthropod hosts, and elicit a variety of phenotypic effects (Figure 1) including feminization, parthenogenesis, male-killing, and sperm-egg incompatibility (cytoplasmic infertility, also referred to as “CI”) (8, 90). Figure 1. A summary of Wolbachia-induced phenotypic effects in arthropods (from Werren et al, 2008) (25). Several studies have shown that the phylogeny of nematode-associated agree closely with the phylogeny of the host species. Filarial nematodes are infected by only two Wolbachia supergroups, C and D(5, 18, 23, 25). This suggests little horizontal movement of Wolbachia among nematode species. In contrast, Wolbachia infections in arthropod hosts show little agreement between host phylogeny and Wolbachia group phylogeny, suggesting extensive horizontal transmission within and between arthropod hosts. Host phenotypic responses, described above as feminization, parthenogenesis, male-killing, and sperm-egg incompatibility, vary depending on the host arthropod species. In other words, a Wolbachia strain that infects one insect taxa, causing CI, may cause male-killing in another insect taxa (1, 2, 5, 18, 23, 25). Casiraghi et al demonstrated that although Wolbachia supergroups are restricted to either arthropods or nematodes, one supergroup was found in both, supporting several theories on horizontal movement of Wolbachia between highly divergent invertebrates (6). These results agree with the results of Bordenstein and Rosengaus (2005), which described a novel supergroup of Wolbachia in Isopterans that is distinct from previously described Wolbachia that infect Isopterans, suggesting horizontal transmission rather than a single acquisition (5). Figure 2 summarizes the phylogeny of Wolbachia supergroups. While the mechanism of Wolbachia horizontal transmission is not well characterized, Kyei-Poku et al. (2005) proposed one mechanism, by which parasitic wasps that feed on other arthropods could acquire a Wolbachia infection and spread it to other uninfected hosts (13). Figure 2. Summary of Wolbachia supergroups (from Werren et al, 2008) (25). The study of Wolbachia in the last fifteen years has revealed several unique traits that previously have not been described in other endosymbiont microbes. Wolbachia has is an exception to the rule of endosymbiosis, as it undergoes extensive recombination, both within and between super-groups,(1, 2, 24, 25) resulting in exceptional levels of diversity among Wolbachia strains. Baldo et al (2006) reported divergence of 6.5 to 9.2% among Wolbachia strains, and have identified 37 unique strains. Why study Wolbachia? In addition to its abilities to extensively recombine between strains, horizontally transmit, and influence host evolution, Wolbachia has become known as the causative agent in pathogenicity of parasitic nematode worms(15). Several diseases, including Lymphatic filariasis, river blindness, chronic inflammatory disease, and elephantitis, are associated with nematode worm infections, and 200 million people worldwide are affected, representing an immense global public health concern (15, 18, 19). While the exact mechanism of Wolbachia’s role in filarial disease has not been fully characterized, data from several studies suggests that Wolbachia is essential to the survival of various parasitic nematode worms and that the immune response to filarial worm infections is likely a response to the release of endo-toxin-like molecules released when worms die or degenerate (15, 18, 21, 25). Antibiotic therapy used in conjunction with traditional anti-filarial treatment has been shown to affect nematode embryonic development, stunt larval development, kill adult worms. Treatment with anti-filarial drugs alone has also been shown to elicit inflammatory response in treated patients, suggesting a Wolbachia-antigen that may be linked to inflammatory disease. These discoveries have led to treatments that decrease host inflammatory disease and ocular inflammation (15, 18, 21, 25). As a result of several recent studies, Wolbachia has been increasingly studied as a target for elimination and treatment of filarial nematode infections as well as treating inflammatory disease in human and livestock hosts. In addition, Wolbachia are currently being studied as agents for controlling pest arthropod populations and insect-borne diseases (15, 16, 18, 21, 25). For example, the vertical transmission of Wolbachia could be used to insert “transgenes”, or foreign genes, into Wolbachia to drive selected traits in an arthropod vector species (16). Alternatively, infected arthropod species could be used to increase cytoplasmic incompatibility in a population, thereby increasing mortality in pest arthropod species (4, 8, 17, 23-26). However, while these mechanisms show promise, reliable manipulation of Wolbachia in this way has yet to be achieved, and controlling filarial nematode populations using Wolbachia has proven more successful. In addition to being instrumental in controlling filarial nematode disease and controlling arthropod populations, the potential for using Wolbachia strains models for understanding evolution seems limitless. Wolbachia may be responsible for a variety of evolutionary mechanisms including host speciation, reproductive isolation, lateral gene transfer from a prokaryotic endosymbiont (Wolbachia) to a eukaryote host, and bacterial recombination (3, 4, 10, 25). Materials and Methods Insect collection Insect samples were collected at random from five different sites using a previouslydescribed sweep-net technique (20, 22). Sampling was conducted for two hours each time and was done both during the morning hours (between 8am and 11am) and during the late afternoon (between 2pm and 5pm). Sampling site information and site descriptions are summarized below: Site: Point Pleasant, NJ Backyard Site Description Suburban neighborhood. Sunny area with lawn grass and long flowering annual and perennial plants. There is very little shade and is very dry. Wall Township, NJ Backyard Damp with maple and oak trees and thick bush. Very little grass grows in the shady area but grass is tall in the sunny areas and large patches of ivy cover most of the uncovered ground. Spring Lake Hts., NJ Backyard Suburban neighborhood. Sunny area with lawn grass and long flowering annual and perennial plants. There is very little shade and is very dry. Wreck Pond, Wall, NJ Riparian area that is part of a watershed that feeds into the Atlantic Ocean. The area is covered with dense low shrubs, pine, maple, and cedar trees, grasses, and sedges. 55-acre State Park that is part of the Manasquan River Estuary that feeds into the Atlantic Ocean. The park size is comoprised of saltmarsh areas and areas of dense low shrubs, pine, maple, and cedar trees, grasses, and sedges. Fisherman’s Cove, Manasquan River, Manasquan, NJ Allaire State Park: 3200 acre park comprised of dense forest composed of oak and maple species. Also includes river floodplain areas and riparian areas. These samples were collected from a wooded area near a small lake near a the main nature center in the park. The ground is sandy and covered in a layer of decaying leaves and other materials. Samples were netted and placed immediately in 95% ethanol then placed in a -20 degree Celcius freezer within twenty minutes of the end of the sampling session. Samples were stored at -20 degrees Celcius in 95% ethanol until sample sorting and washing. Insect samples were sorted and washed using the MBL/Discover the Microbes Within insect identification lab protocol (7). Samples were sorted into various morphospecies (Table 1) per the MBL/Discover the Microbes Within insect identification lab protocol and were processed rapidly to maintain DNA quality. Redundancies and cross-referenced species were noted, where applicable and species identification was made, when possible. Furthermore, voucher species were preserved frozen in 95% ethanol for future species identification. Sorted samples were placed back in the -20 degree Celcius freezer until DNA extraction was performed. DNA Extraction DNA extractions were performed from a minimum of one and a maximum of eleven samples from each representative morphospecies per the MBL/Discover the Microbes Within DNA Extraction protocol and the Qiagen DNeasy tissue DNA extraction protocol (7). Samples were chosen at random for DNA extraction. If the insect measured larger than 2-5mm, a 2-3mm section was removed from the insect abdomen for DNA extraction. Otherwise, the entire insect was used for DNA extraction. Samples were completely ground/pestled, per the Qiagen DNeasy protocol and were processed exactly according to the protocol to ensure DNA quality. Genomic DNA samples were run on 1% agarose gels for the first two sets of DNA extractions to qualitatively test for DNA presence and quality. PCR Amplification and Gel Electrophoresis The MBL/Discover The Microbes Within PCR protocol was used to amplify a Wolbachiaspecific 438 base-pair fragment (WSPEC) of the prokaryotic 16SrRNA gene and a 658 base-pair fragment of the eukaryotic cytochrome oxidase (CO) gene (7). PCR was carried out in 25µl volumes using the GE-Healthcare PCR bead kit and the MBL/Discover The Microbes Within PCR protocol. Three controls were used for each set of samples amplified. The controls are summarized below: (+) Nasonia – a known Wolbachia-positive sample (-) Nasonia – a known Wolbachia-negative sample No-DNA – a sterile-milliQ water negative control. PCR was conducted using the following primers: WSPEC Forward: (5’-CATACCTATTCGAAGGGATAG-3’) WSPEC Reverse: (5’-AGCTTCGAGTGAAACCAATTC-3’) CO Forward (are LCO1490): (5’-GGTCAACAAATCATAAAGATATTGG-3’) CO Reverse (HCO2198): (5’-TAAACTTCAGGGTGACCAAAAAATCA-3’) PCR cycle information is summarized below: 1 cycle 1 min @ 94 C 38 Cycles 1 min @ 94 C 1.5 min @ 45 C 1 min @ 72 C 1 cycle 5 min. @72 C Each PCR product was run on 1% agarose using 1X TAE buffer. 10µl of each PCR product was electrophoresed for 48 minutes at 75 volts in a chilled gel-boat. PCR product size was determined using a 1kb ladder. Gels were stained with ethidium bromide and then visualized in UV light. PCR products were frozen immediately after PCR cycling was completed to ensure DNA quality. Positive Wolbachia infections were identified and recorded in the results section. Results A total of 96 unique insect samples were PCR tested for both the eukaryote CO gene fragment and the prokaryote Wolbachia-specific 16SrRNA fragment. 29% of all samples tested were positive for the Wolbachia-specific fragment, in close agreement with previous estimates (10, 23-25) PCR results are summarized below: Table 1: Morphospecies list and PCR test results. Positive results indicate a positive Wolbachiaspecific 16SrRNA fragment. Morphospecies FB2010-17 Order Hymenoptera FB2010-21-1 Hymenoptera FB2010-21-2 Hymenoptera FB2010-22-1 Hymenoptera FB2010-22-1 dup pcr Hymenoptera FB2010-22-2 Hymenoptera FB2010-22-2 dup pcr Hymenoptera Date collected 7/19/10 7/20/10 7/20/10 7/20/10 7/20/10 7/20/10 7/20/10 FB2010-26-1 Diptera 7/22/10 FB2010-26-2 Diptera 7/22/10 FB2010-28 Hemiptera 7/22/10 FB2010-28 dup pcr Hemiptera 7/22/10 FB2010-31 Orthoptera 7/22/10 FB2010-33 Diptera 7/22/10 FB2010-34-1 Hemiptera 7/22/10 FB2010-34-2 hemiptera 7/22/10 FB2010-36 Hemiptera 7/22/10 FB2010-39 Hymenoptera 7/22/10 FB2010-39 dup pcr Hymenoptera 7/22/10 FB2010-40-1 Hemiptera 7/22/10 FB2010-40-2 Hemiptera 7/22/10 FB2010-41-1 Diptera 7/22/10 FB2010-41-2 Diptera 7/22/10 FB2010-42 Diptera 7/22/10 FB2010-46-1 Hymenoptera 7/22/10 FB2010-46-1 dup pcr Hymenoptera 7/22/10 FB2010-46-2 Hymenoptera 7/22/10 FB2010-47 FB2010-47 Hymenoptera Hymenoptera 7/22/10 7/22/10 Location Pt. Pleasant Allaire State Park Allaire State Park Allaire State Park Allaire State Park Allaire State Park Allaire State Park Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point PCR test + + + + Species/Description JDN2010-27-1 Hymenoptera 7/20/10 JDN2010-27-2 Hymenoptera 7/20/10 Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Point Pleasant Allaire State Park Allaire State Park Allaire State Park Allaire State Park Allaire State Park Allaire State Park Allaire State Park Allaire State Park Allaire State Park Allaire State Park Spring Lake Hts Spring Lake Hts JDN2010-29-1 Coleoptera 7/20/10 Wreck Pond Tiny brown beetle JDN2010-29-2 Coleoptera 7/20/10 Wreck Pond Tiny brown beetle JDN2010-29-3 Coleoptera 7/20/10 Wreck Pond Tiny brown beetle JDN2010-29-4 Coleoptera 7/20/10 Wreck Pond Tiny brown beetle JDN2010-31-1 Hymenoptera 7/20/10 Wreck Pond Spiny black ants JDN2010-31-2 Hymenoptera 7/20/10 Wreck Pond Spiny black ants JDN2010-31-3 Hymenoptera 7/20/10 Wreck Pond Spiny black ants JDN2010-31-4 Hymenoptera 7/20/10 Wreck Pond Spiny black ants JDN2010-32 Coleoptera 7/20/10 Wreck Pond smallish brown round beetle JDN2010-33 Diptera 7/20/10 Wreck Pond zebra wing fly JDN2010-34 Coleoptera 7/21/10 Wreck Pond FB2010-48-1 Hemiptera 7/22/10 FB2010-48-2 Hemiptera 7/22/10 FB2010-51 Hymenoptera 7/22/10 FB2010-52 Hymenoptera 7/22/10 FB2010-53 Hymenoptera 7/22/10 FB2010-54-1 Hemiptera 7/22/10 7/23/2010 FB2010-54-1 dup pcr Hemiptera FB2010-54-2 Hemiptera FB2010-54-2 dup pcr Hemiptera FB2010-54-3 Hemiptera FB2010-54-3 dup pcr Hemiptera FB2010-54-4 Hemiptera FB2010-54-4 dup pcr Hemiptera 7/23/2010 7/23/2010 7/23/2010 7/23/2010 7/23/2010 7/23/2010 7/23/2010 FB2010-56 Hymenoptera 7/23/2010 FB2010-58 Diptera FB2010-7 Hemiptera 7/23/2010 very weak + + + + + medium black beetle JDN2010-35-1 Diptera 7/21/10 Wreck Pond + zebra wing fly JDN2010-35-2 Diptera 7/21/10 Wreck Pond + zebra wing fly JDN2010-35-3 Diptera 7/21/10 Wreck Pond JDN2010-35-4 Diptera 7/21/10 Wreck Pond JDN2010-36-1 Diptera 7/21/10 Wreck Pond longtail zebra wing fly JDN2010-36-2 Diptera 7/21/10 Wreck Pond longtail zebra wing fly JDN2010-37 Hemiptera 7/21/10 Wreck Pond Graphocephala versuta JDN2010-38 Coleoptera 7/21/10 Wreck Pond + JDN2010-39 Coleoptera 7/21/10 Wreck Pond + JDN2010-40-1 Hemiptera 7/21/10 Wreck Pond zebra face bug JDN2010-40-2 Hemiptera 7/21/10 Wreck Pond JDN2010-41-1 Coleoptera 7/21/10 Wreck Pond JDN2010-41-2 Coleoptera 7/21/10 Wreck Pond JDN2010-41-3 Coleoptera 7/21/10 Wreck Pond zebra face bug translucent wing, tiger striped beetle translucent wing, tiger striped beetle translucent wing, tiger striped beetle JDN2010-42 Hemiptera 7/21/10 JDN2010-47 Coleoptera 7/21/10 JDN2010-48 Hymenoptera 7/21/10 JDN2010-51 Coleoptera 7/21/10 JDN2010-55 Hymenoptera 7/21/10 JDN2010-61-1 Hymenoptera 7/22/10 JDN2010-61-2 Hymenoptera 7/22/10 JDN2010-61-3 Hymenoptera 7/22/10 JDN2010-61-4 Hymenoptera 7/22/10 JDN2010-61-5 Hymenoptera 7/22/10 JDN2010-61-6 Hymenoptera 7/22/10 JDN2010-62 Hymenoptera 7/22/10 JDN2010-63 Hymenoptera 7/22/10 JDN2010-66 Hymenoptera 7/22/10 JDN2010-67 Diptera 7/22/10 Wreck Pond Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove zebra wing fly + zebra wing fly + calico-checker tail bug + Harmonia axyridis + Dark green/blue wasp medium black beetle (same as morpho 34) + + + + + black/green small wasp black hornet, 2 yellow V's on face, 2 yellow spots on shoulder black hornet, 2 yellow V's on face, 2 yellow spots on shoulder black hornet, 2 yellow V's on face, 2 yellow spots on shoulder black hornet, 2 yellow V's on face, 2 yellow spots on shoulder black hornet, 2 yellow V's on face, 2 yellow spots on shoulder black hornet, 2 yellow V's on face, 2 yellow spots on shoulder black hornet, 2 yellow V's on face, 1 yellow dot on shoulder black hornet, 2 white V's on face, 2 white dots on shoulder small, dark green/black hornet, long stinger zebra wing fly JDN2010-70-1 Coleoptera 7/22/10 JDN2010-70-10 Coleoptera 7/22/10 JDN2010-70-11 Coleoptera 7/22/10 JDN2010-70-2 Coleoptera 7/22/10 JDN2010-70-3 Coleoptera 7/22/10 JDN2010-70-4 Coleoptera 7/22/10 JDN2010-70-5 Coleoptera 7/22/10 JDN2010-70-6 Coleoptera 7/22/10 JDN2010-70-7 Coleoptera 7/22/10 JDN2010-70-8 Coleoptera 7/22/10 JDN2010-70-9 Coleoptera 7/22/10 Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Manasquan Cove Small silver beetle + Small silver beetle + Small silver beetle + Small silver beetle + Small silver beetle + Small silver beetle + Small silver beetle + Small silver beetle Small silver beetle Small silver beetle Small silver beetle Figure 3. Proportion of PCR positives as a function of the total number of insects tested (n=96). Figure 4. Proportion of PCR positives represented by each insect order. Figure 5. Insect genomic DNA gels Figure 6. PCR gel results. Conclusion According to previous data the expected amount of insects infected with Wolbachia is around 20% and that about 60% of individuals in a species would be infected. We hypothesized that the amount of our infected samples would be around the same percentage. After collecting samples and running pcr test we found that out of the 96 individual samples tested that approximately 30% were infected with Wolbachia. Out of the morpho species that were infected, up to 86% of individuals were infected. These percentages were higher then expected percent, however we were not able to conduct PCR testing all individual samples collected. In order to test this hypothesize farther we would need to collect and test a larger number of samples and test more replicates per species. . In addition we also asked if the location affected the amount of infections. We saw that certain areas did have more infections then others but we would need to do more testing to further characterize local ecological effects. References 1. Baldo L., Bordenstein S., Wernegreen J. J., and Werren J. H. 2006. Widespread recombination throughout Wolbachia genomes. U.S. Patent 2. Mol Biol Evol. United States 23:437-49. 2. Baldo L., Dunning Hotopp J. C., Jolley K. A., Bordenstein S. R., Biber S. A., Choudhury R. R., Hayashi C., Maiden M. C., Tettelin H., and Werren J. H. 2006. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. U.S. Patent 11. Appl Environ Microbiol. United States 72:7098-110. 3. Bordenstein S. R., and Reznikoff W. S. 2005. Mobile DNA in obligate intracellular bacteria. U.S. Patent 9. Nat Rev Microbiol. England 3:688-99. 4. Bordenstein S. R., and Werren J. H. 2007. Bidirectional incompatibility among divergent Wolbachia and incompatibility level differences among closely related Wolbachia in Nasonia. U.S. Patent 3. Heredity. England 99:278-87. 5. Bordenstein S., and Rosengaus R. B. 2005. Discovery of a novel Wolbachia super group in Isoptera. U.S. Patent 6. Curr Microbiol. United States 51:393-8. 6. Casiraghi M., Bordenstein S. R., Baldo L., Lo N., Beninati T., Wernegreen J. J., Werren J. H., and Bandi C. 2005. Phylogeny of Wolbachia pipientis based on gltA, groEL and ftsZ gene sequences: clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. U.S. Patent Pt 12. Microbiology. England 151:401522. 7. Discover the Microbes Within: The Wolbachia Project Lab Series . Marine Biological Labs 8. Dobson S. L., Fox C. W., and Jiggins F. M. 2002. The effect of Wolbachia-induced cytoplasmic incompatibility on host population size in natural and manipulated systems. U.S. Patent 1490. Proc Biol Sci. England 269:437-45. 9. Hertig M., and Wolbach S. B. 1924. Studies on Rickettsia-Like Micro-Organisms in Insects. U.S. Patent 3. J Med Res. United States 44:329-374.7. 10. Hilgenboecker K., Hammerstein P., Schlattmann P., Telschow A., and Werren J. H. 2008. How many species are infected with Wolbachia?--A statistical analysis of current data. U.S. Patent 2. FEMS Microbiol Lett. England 281:215-20. 11. Ishmael N., Hotopp J. C., Ioannidis P., Biber S., Sakamoto J., Siozios S., Nene V., Werren J., Bourtzis K., Bordenstein S. R., and Tettelin H. 2009. Extensive genomic diversity of closely related Wolbachia strains. U.S. Patent Pt 7. Microbiology. England 155:2211-22. 12. Kent B. N., and Bordenstein S. R. 2010. Phage WO of Wolbachia: lambda of the endosymbiont world. Trends in Microbiology. Elsevier. 13. Kyei-Poku G. K., Colwell D. D., Coghlin P., Benkel B., and Floate K. D. 2005. On the ubiquity and phylogeny of Wolbachia in lice. U.S. Patent 1. Mol Ecol. England 14:285-94. 14. Lo N., Paraskevopoulos C., Bourtzis K., O'Neill S. L., Werren J. H., Bordenstein S. R., and Bandi C. 2007. Taxonomic status of the intracellular bacterium Wolbachia pipientis. U.S. Patent Pt 3. Int J Syst Evol Microbiol. England 57:654-7. 15. Pfarr K. M., and Hoerauf A. M. 2006. Antibiotics which target the Wolbachia endosymbionts of filarial parasites: a new strategy for control of filariasis and amelioration of pathology. U.S. Patent 2. Mini Rev Med Chem. Netherlands 6:203-10. 16. Sinkins S. P., and Godfray H. C. 2004. Use of Wolbachia to drive nuclear transgenes through insect populations. U.S. Patent 1546. Proc Biol Sci. England 271:1421-6. 17. Sinkins S. P., and Gould F. 2006. Gene drive systems for insect disease vectors. U.S. Patent 6. Nat Rev Genet. England 7:427-35. 18. Taylor M. J. 2002. Wolbachia endosymbiotic bacteria of filarial nematodes. A new insight into disease pathogenesis and control. U.S. Patent 4. Archives of Medical Research. Elsevier 33:422-424. 19. Taylor M. J., Bandi C., Hoerauf A. M., and Lazdins J. 2000. Wolbachia bacteria of filarial nematodes: a target for control?. U.S. Patent 5. Parasitology Today. Amsterdam: Elsevier Science Publishers, c1985-c2000. 16:179-180. 20. Thipaksorn A., Jamnongluk W., and Kittayapong P. 2003. Molecular evidence of Wolbachia infection in natural populations of tropical odonates. U.S. Patent 4. Curr Microbiol. United States 47:314-8. 21. Turner J. D., Mand S., Debrah A. Y., Muehlfeld J., Pfarr K., McGarry H. F., Adjei O., Taylor M. J., and Hoerauf A. 2006. A randomized, double-blind clinical trial of a 3-week course of doxycycline plus albendazole and ivermectin for the treatment of Wuchereria bancrofti infection. U.S. Patent 8. Clin Infect Dis. United States 42:1081-9. 22. Unckless R. L., Boelio L. M., Herren J. K., and Jaenike J. 2009. Wolbachia as populations within individual insects: causes and consequences of density variation in natural populations. U.S. Patent 1668. Proc Biol Sci. England 276:2805-11. 23. Werren J. H. 1997. Biology of wolbachia. U.S. Patent 1. Annual Review of Entomology. Annual Reviews 42:587-609. 24. Werren J. H., and Windsor D. M. 2000. Wolbachia infection frequencies in insects: evidence of a global equilibrium?. U.S. Patent 1450. Proceedings of the Royal Society B: Biological Sciences. The Royal Society 267:1277. 26. Zabalou S., Riegler M., Theodorakopoulou M., Stauffer C., Savakis C., and Bourtzis K. 2004. Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. U.S. Patent 42. Proceedings of the National Academy of Sciences of the United States of America. National Acad Sciences 101:15042.