View/Open
advertisement

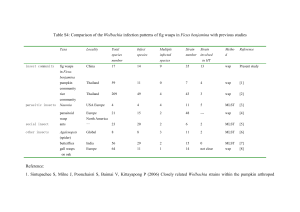
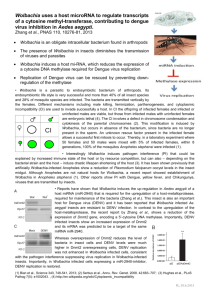
Vector competence is a complex characteristic which governs an insect’s ability to acquire, support the development and transmit a parasite from one host to another. It influences variation in disease transmission among mosquito populations, hence affecting disease epidemiology. In this project, I have studied some aspects of ecological interactions and genetic factors in a step towards understanding how these affect variation in disease transmission and exploiting these in future disease control programmes. Mosquito gut bacteria affect the development of parasites ingested by mosquitoes. As different bacterial species have different effects, dissimilarities in gut composition could be an important cause of variation in vector populations. The first study investigates the gut microbiome of mosquitoes collected from Kenya. Using 454 pyrosequencing of 16S rRNA, I provide a comprehensive catalogue of the gut composition of 8 species of mosquitoes (Chapter 2). I show that while there is greater variation within host species (fixation index= 0.64), different mosquito species tend to have rather similar gut bacteria. An individual mosquito gut has a low diversity of bacteria with, the microbiota being dominated by a single Operational Taxonomic Unit. This suggests that gut bacteria may be one factor influencing within-species variation in disease transmission, and a minor factor in between-species variation. Wolbachia endosymbionts are able to reduce the intensity and development of RNA viruses and metazoan parasites in their insect hosts, blocking the transmission of such parasites. This makes Wolbachia a likely candidate for control programmes. I extend the investigation of naturally-occurring bacteria to Wolbachia (Chapter 3). Using the gut samples used in Chapter 2, I amplify the Wolbachia surface protein gene to identify Wolbachia infections. I identify Wolbachia in Aedes bromeliae, a vector of yellow fever, and its close relative Aedes metallicus and in Mansonia uniformis and Mansonia africana, which are competent vectors of human bancroftian filariasis. Aedes bromeliae showed the highest prevalence (75%) suggesting that this strain of Wolbachia may be manipulating the host reproduction by cytoplasmic incompatibility. Using a multi locus typing system and accounting for effects of recombination in the construction of bacterial phylogeny, I show that these mosquito Wolbachia strains cluster into supergroups A and B of Wolbachia. The phylogeny also shows significant recombination events indicating horizontal transfer events between taxa. These Wolbachia strains, isolated from the disease vectors, may be reducing parasite intensity and transmission, and could be a better choice for transinfecting other mosquito vectors rather than distantly related strains. Previous studies show that high frequency of susceptibility to Brugia pahangi exists among populations of Aedes aegypti from East Africa, providing an excellent resource for investigating variation in a natural population. I test the frequency of susceptibility of peri-domestic subpopulations of Aedes aegypti collected from Kenya to Brugia malayi (Chapter 4). The results are consistent with previous data with up to 30% of individuals being susceptible. The number of susceptible individuals varied significantly between populations (Fisher‘s exact: p= 0.03). These populations now provide the resource to identify polymorphisms associated with susceptibility to Brugia and also enable comparison with results obtained from laboratory strains. In Chapter 5, I continue with efforts to identify and map quantitative trait loci (QTL) associated with Brugia susceptibility in Aedes aegypti. However, with the Aedes genome still highly fragmented with many supercontigs having no chromosomal assignments, mapping the gene to a definitive locus is almost impossible. Using an improved DNAbased mapping technology, Restricted-site Associated DNA tags (RADtags), I make novel assignments of 79 supercontigs to the 3 chromosomes of Aedes aegypti. These new assignments account for 122Mb of the genome, increasing the percentage genome mapped to ≈40%. The technique also identifies potential scaffold misassemblies and misassignments of supercontigs to chromosomes. I also use the same method to prepare libraries for sequencing which will provide more markers and allow mapping and identification of candidate genes which can be evaluated for involvement in susceptibility to Brugia infections. Aedes aegypti and Anopheles gambiae share similarities in their immune proteins, but little is known about the functions of immune proteins in Aedes aegypti. To be able to make functional comparisons between mosquito vectors, I inoculate Sephadex beads into a laboratory strain of Aedes aegypti to investigate the expression of pathogen recognition genes (Chapter 6). Thioester-containing proteins (TEPs) show significant up-regulation (p= 0.03-0.0002) with up to 7-fold increase in gene expression of TEP20 in immunechallenged individuals compared to non-challenged controls. TEP20 is an orthologue of Anopheles gambiae TEP1, emphasising the evolutionary function of TEPs in immune activation. As TEP1 is an important determinant of vectorial capacity in Anopheles gambiae, this indicates that TEPs may also be an important factor influencing variation in susceptibility to pathogens in Aedes aegypti. Generally, this project has contributed to three broad areas of factors that influence variability in diseases transmission by mosquitoes: ecological interactions with bacteria, host genetic background and immune system. The results, resources and techniques used in this thesis can be widely used in further studies in these areas and extended to other mosquito vectors and natural populations.