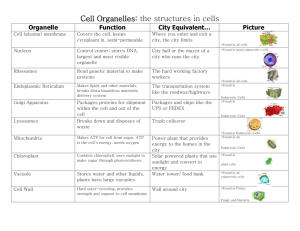
Hartmannella vermiformis 598 - Hv Contig 598 is removed due to
advertisement

Hartmannella vermiformis 598 - Hv Contig 598 is removed due to the hit to a Jakobid sequence in PEPdb. 863 - Despite the presence of multiple eukaryotic sequences in the tree, this sequence passes as an LGT due to strong clustering with actinobacterial sequence and high levels of bootstrap support for separation from the other eukaryotic sequences. 918 - This clone is a strong pass – there is only one actinobacterial sequence similar to it, and no other eukaryotic hits in any database anywere. 465 - This is a strong positive hit, with no eukaryotic orthologs or paralogs and strong bootstrap clustering with bacterial taxa. 59 - There are some eukaryotic hits, but the Hartmannella sequence clusters strongly apart on a bacterial branch. 664 - There is a single eukaryotic hit to an EST from Sorghum. The identity is sufficiently strong that we regard it as a likely case of amoebal contamination in the EST library. Apart from that there are no eukaryotic hits. 1187 - Eliminated as a lateral candidate due to the presence of clear Fungal orthologs. Nonetheless this could be a basal (to Amoebozoa+Fungi) LGT for an anaerobic-linked nitrite reductase. However there may be poorly supported clustering with plant paralogs as well. 277 - Cluster 277 hits only three bacterial sequences and no eukaryotic sequences. It is thus a solid candidate for an LGT event. 1030 - Pinus has a perfect match hit to a single EST – once again we are inclined to classify this as a contaminant due to the excessively high level of sequence similarity. There appear to be plant paralogs to this gene, but they are not strongly similar to the Amoebozoan sequences. This is nonetheless a very questionable candidate even based upon bootstraps in the absence of the additional EST. 474 - There are a large number of eukaryotic hits to this sequence in both Genbank and PEPdb. We therefore reject it at the secondary screening. 480 - This sequence is a strong pass. It clearly clusters with cyanobacterial sequence and has no obvious eukaryotic orthologs or paralogs. 102 - This sequence is another strong pass as it clusters strictly with bacterial genes. 891 - This sequence is eliminated – multiple databases contain multiple eukaryotic genes which have substantial sequence similarity. 946 - Fructokinase. This sequence is eliminated on secondary screening due to the presence of multiple eukaryotic sequences with strong similarity. 143 - This sequence is eliminated on tertiary screening due to poor bootstrap support for basal nodes, with possible linkage to other eukaryotic sequences. 1198 - Rejected on tertiary screening – not strongly separated from the Arabidopsis sequence. 65 - Reject due to incomplete seperation from Giardia/Spironucleus sequences on tertiary screening (poor bootstraps). 445 - Rejected on tertiary screening – clearly clusters with other eukaryotic sequences. 486 - Rejected on secondary screen due to hits against multiple eukaryotes in multiple databases. 496 - No eukaryotic hits, tertiary screening clusters it clearly with proteobacteria. Passes as LGT candidate. 504 - Rejected on secondary screen due to presence of multiple eukarotic hits. 524 - Rejected on secondary screen due to presence of multiple eukarotic hits. 584 - Clusters at 63% bootstraps with with spirochaete sequence. Appears to be cleanly separate from other eukaryotic sequences. 615 - Rejected on secondary screen due to presence of many eukarotic hits. 627 - Clusters strongly with Lactobacillus, Streptococcus, Burkholderia. 733 - Rejected on tertiary screen due to insufficient separation from other eukaryotic sequences (poor bootstraps). 751 - Rejected on secondary screen due to multiple eukaryotic hits in other databases. 782 - Rejected on secondary screen due to multiple eukaryotic hits in other databases. 1048 - Rejected on secondary screen due to multiple eukaryotic his in other databases. 1091 - Strong clustering with cyanobacterial sequences, no apparent eukaryotic sequences in any database. Toxoplasma 837 - Eliminated on secondary screen due to many eukaryotic hits outside of the Apicomplexa. 425 - Eliminated on secondary screen due to many eukaryotic hits outside of the Apicomplexa. 1194 - Eliminated on secondary screen due to many eukaryotic hits outside of the Apicomplexa. Chlamydomonas 391 - Eliminated on secondary screen due to many eukaryotic hits outside of the Chlorophyta. 648 - Eliminated on secondary screen due to multiple eukaryotic hits outside of the Chlorophyta. 727 - Eliminated on secondary screen due to many eukaryotic hits outside of the Chlorophyta. 611 - Eliminated on secondary screen due to multiple eukaryotic hits outside of the Chlorophyta. 854 - Eliminated on tertiary screen due to strong clustering with the cyanobacteria. Drosophila 74 - Eliminated on tertiary screening – does not strongly separate from other eukaryotic sequences. MUST DO PROPER BOOTS 195 - Eliminated on secondary screen due to many eukaryotic hits outside of the Metazoa. 645 - Passes tertiary screen as a candidate LGT since there are no hits to eukaryotic phyla apart from Metazoa. 948 - Eliminated on secondary screen due to many eukaryotic hits outside of the Metazoa. 1462 - Passes tertiary screen as a candidate LGT since there are no hits to eukaryotic phyla apart from Metazoa. 1497 - This is essentially an identical sequence to 1462 – may have an unprocessed intron. 1499 - Eliminated on secondary screen due to many eukaryotic hits outside of the Metazoa. 620 - Eliminated on secondary screen due to eukaryotic hits outside of the Metazoa. 876 - Eliminated on secondary screen due to eukaryotic hits outside of the Metazoa. 1174 - Eliminated on secondary screen due to many eukaryotic hits outside of the Metazoa. 1397 - Eliminated on secondary screen due to many eukaryotic hits outside of the Metazoa. 153 - Eliminated on tertiary screen due to clustering with additional eukaryotic taxa beyond the Metazoa. Dictyostelium AF 275 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 303 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 382 - Eliminated on tertiary screen due to clustering with Jakoba libera. 1031 - Reject on secondary and tertiary screen due to clustering with eukaryotes outside of Amoebozoa. This is on the other hand one of the LGT candidates which could tie Amoebozoa with a subset of the Excavates and must be looked at carefully (HV598). Reject at current stringent screening level, but keep in the larger queue. 235 - Rejected due to hits to Metazoa. 548 - Hits only Microbulbifer degradans. Hits to some other Amoebozoans as well. Very distant (e-05) eukaryotic hits are so divergent as to be unalignable. Microbulbifer also an extremely poor alignment. Inclined to dismiss, but accept on a technical level. 685 - No hits to any eukaryotic sequences. candidate. Passed as an LGT 737 - No hits to any eukaryotes outside of the Amoebozoa. Passed as an LGT candidate. 816 - Rejected due to clustering with additional eukaryotic taxa outside the Amoebozoa. Dictyostelium VF 151 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 196 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 227 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 228 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 502 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 613 - Eliminated on tertiary screen due to clustering with eukaryotic taxa apart from the Amoebozoa. 781 - Thymidylate synthase. This candidate has been rejected due to clustering with Reclinomonas americana. 792 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 890 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 997 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 998 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 18 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 138 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 208 - Eliminated on tertiary screen due to clustering with eukaryotic taxa apart from the Amoebozoa. 429 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 456 - Eliminated on tertiary screen due to clustering with eukaryotic taxa apart from the Amoebozoa. 707 - Ornithine/Arginine decarboxylase. Dictyostelium with Malawimonas. Appears to cluster 842 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 984 - Eliminated on tertiary screen due to clustering with eukaryotic taxa apart from the Amoebozoa. 999 - Glycosyl hydrolase but not HV598. Appears to cluster Dictyostelium with Acanthamoeba and Trimastix. Acanthamoeba 216 - cmfA-like. Appears to cluster Dictyostelium with Acanthamoeba, Hartmannella, and Malawimonas. 233 - Eliminated on tertiary screen due to clustering with eukaryotic taxa apart from the Amoebozoa. 367 - Eliminated on tertiary screen due to clustering with eukaryotic taxa apart from the Amoebozoa. 452 - Appears to cluster strongly away from other Eukaryotic paralogs. This sequence therefore passes as an LGT candidate. 565 - Eliminated on tertiary screen due to clustering with eukaryotic taxa apart from the Amoebozoa. 705 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 832 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 896 - Eliminated on tertiary screen due to possible clustering with broad eukaryotic radiation. Bootstraps are unconvincing. 1020 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 1161 - Hits a Micromonas cluster in PEPdb. secondary screen. Eliminated on 114 - Eliminated on tertiary screen due to clustering with eukaryotic taxa apart from the Amoebozoa. 115 - Eliminated on tertiary screen due to possible clustering with eukaryotic taxa apart from the Amoebozoa. 434 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 559 - Eliminated on tertiary screen due to clustering with eukaryotic taxa apart from the Amoebozoa. 630 - Eliminated on tertiary screen due to clustering with eukaryotic taxa apart from the Amoebozoa. 850 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 971 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 977 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 1002 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 1138 - Eliminated on tertiary screen due to clustering with eukaryotic taxa apart from the Amoebozoa. 1140 - Only hits prokaryotic sequences. LGT. Passes as a candidate 1155 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa. 1199 - Eliminated on tertiary screen due to clustering with eukaryotic taxa apart from the Amoebozoa. 1203 - Only hits prokaryotic sequences. LGT. Passes as a candidate 1287 - Eliminated on secondary screen due to eukaryotic hits outside the Amoebozoa.