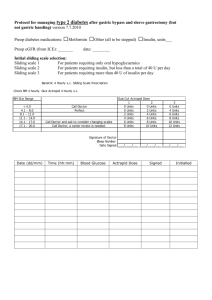
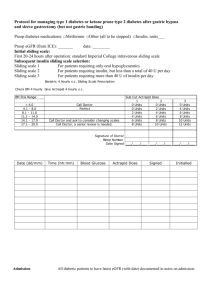
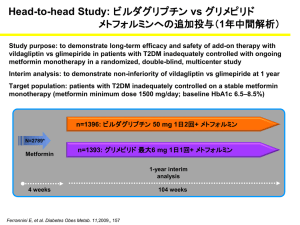
Supplementary Data (doc 33K)
advertisement

Supplemental Datas Metformin causes an early cell cycle arrest in G0/G1 phase Inhibition in proliferation would indicate an anomaly in the cell cycle. To examine this, A375 cells were treated with metformin for 24h, and phases of cell cycle were analyzed by flow cytometry. As shown in the figure S1A, 5mM and 10mM metformin treatment led to accumulation of 6% and 8% in G0/G1 phase with corresponding decrease in percentage of G2 phase fraction as compared with control, PBS treated cells. In this condition, metformin did not induce a sub G1 accumulation suggestive of apoptosis. To confirm this result, we performed cell cycle synchronization experiment; By using a double thymidine block protocol, 98% of the cells were effectively arrested at the G1/S phase of the cell cycle (data not shown). At the time of release, cells were stimulated with 10 mM metformin. As previously, cell cycle analysis revealed that 24 h of metformin treatment resulted in accumulation of cells in G0/G1 phase (18%), with no increase of cells detected in Sub G1 phase (Fig. S1B). By western blot, we examined key proteins implicated in the G0/G1 phase transition. Consistent with the observed G0/G1 cell cycle arrest, metformin reduced expression of cyclin D1 and a concomitant dephosphorylation of pRb. We observed an increase in cyclin-dependent kinase inhibitor p21 since there was no changes in the expression of p27 (Fig. S1C). Early cell cycle arrest in G0/G1 phase is involved in autophagy mediated by metformin Early cell cycle arrest in G0/G1 phase mediated by metformin is characterized by an increase of p21 expression. So, to determine whether the initial G0/G1 cell cycle arrest contributes to autophagy, we prevent metformin induced p21 increase by siRNA p21. Western blot presented on Fig S1D shows that p21 siRNA abolished the increase of p21 expression and decreased the expression and conversion of LC3-b induced by metformin. These results suggest that initial cell cycle arrest induced by metformin is involved in autophagy. Autophagy mediated by metformin is involved in apoptosis Using ATG5 siRNA, we obtained the same results that LC3 siRNA. Indeed, ATG5 siRNA, which drastically reduced the expression of the protein, prevented also PARP cleavage and cell death induced by metformin (Fig. S4 A and B). The apoptosis inhibitor, QVD-OPH was capable to inhibit PARP cleavage and cell death induced by metformin. In contrast, Q-VD-OPH failed to reduce LC3-II expression-mediated by metformin. As expected, combination of both ATG5 siRNA and Q-VD-OPH abolished autophagy, apoptosis and cell death triggered by metformin. These results confirm the fact that apoptosis is a consequence of autophagy induced by metformin. In our previous experiments, inhibition of apoptosis was mediated by specific caspase inhibitor, Q-VD-OPH. Further, we have performed new experiments in which apoptosis was inhibited by caspase 3 siRNA. As show in Fig.S4 C, inhibition of caspase 3 by siRNA inhibited apoptosis induced by metformin but not autophagy confirming results obtained with Q-VD-OPH. Finally, Beclin 1 siRNA, which drastically reduced the expression of the protein, is unable to prevented PARP cleavage mediated by metformin (Fig. S4D). As expected, Q-VDOPH was capable to inhibit PARP cleavage induced by metformin. In contrast, Q-VD-OPH failed to impair LC3-II accumulation-mediated by metformin. In addition, Beclin 1 siRNA is unable to prevented LC3-b conversion induced by metformin and combination of both Beclin 1 siRNA and Q-VD-OPH did not abolished autophagy triggered by metformin. Legend Figures Figure S1. Metformin blocks cell cycle in G0/G1 and affects the expression level of the cell cycle proteins. (a) Cells treated for 24 hours with indicated concentration of metformin were detached and their DNA contents were measured by flow cytometry. The table displays the percentage of cells in G0/G1, S and sub-G1 stages (apoptotic cells). One representative experiment of three is shown. (b) Flow cytometry analysis of cells treated during 24h with 10mM of metformin. synchronized at the onset of S phase by double thymidine block (c) A375 melanoma cells were treated for 24 hours with increasing concentrations of metformin. Cell lysates were separated by SDS–PAGE and analyzed by western blot using the indicated antibody. HSP60 was used as a loading control. One representative experiment of three is shown. (d) A375 melanoma cells transfected with p21 siRNA were treated for 72 hours with 10 mM metformin. Cell lysates were separated by SDS–PAGE and analyzed by western blot using the indicated antibody. HSP60 was used as a loading control. One representative experiment of three is shown. Figure S2. Metformin triggers autophagy in melanoma SKMel28 cells (a) Cells were treated with 10 mM of metformin for 72h. Electronic microscopy images presenting ultrastructure in representative control (PBS) and metformin treated melanoma cells. Arrowheads, autophagosomes. Nu, nucleus; M, mitochondria. Figure S3. Metformin induces LC3b conversion and PARP cleavages in A375 cells A375 cells were incubated with metformin (10mM) for the indicated times. Cell lysates were separated by SDS-PAGE and analysed by western blot using the indicated antibody. HSP60 was used as loading control. One representative experiment of three is shown. Figure S4. Autophagy is involved in apoptosis and antiproliferative effects of metformin. (a) A375 cells were transfected with either control or ATG5-specific siRNAs. Cells were next preincubated with or without Q-VD-OPh (20 µM) and exposed to metformin (10 mM) for 96h. LC3-b expression and PARP cleavage were analyzed by western blotting. HSP60 was used as a loading control. (b) FACS analysis using propidium iodide was used to determine cell death. The results are expressed as percentage of cell death compared with control. Data are expressed as percentage of apoptotic cells and represent the mean ± SD of 3 independent experiments. (c) A375 cells were transfected with either control, LC3-specific and/or caspase 3-specific siRNAs. Cells were next exposed to metformin (10 mM) for 96h. LC3-b expression and PARP cleavage were analyzed by western blotting. HSP60 was used as a loading control. One representative experiment of three is shown. d) A375 cells were transfected with either control or Beclin 1-specific siRNAs. Cells were next preincubated with or without Q-VD-OPh (20 µM) and exposed to metformin (10 mM) for 96h. LC3-b expression and PARP cleavage were analyzed by western blotting. HSP60 was used as a loading control.