A 3D Immersive Virtual Environment for Secondary Biology
advertisement

A 3D Immersive Virtual Environment for Secondary Biology Education
Otto Borchert, Guy Hokanson, Brian M. Slator, Bradley Vender, Peng Yan, Vaibhav Aggarwal, Matti Kariluoma
Department of Computer Science and Operations Research
North Dakota State University
United States
{otto.borchert, guy.hokanson, brian.slator, bradley.vender, peng.yan, vaibhav.aggarwal,
matti.m.kariluoma}@ndsu.edu
Andrew “Mazz” Marry
Department of Biosciences
Minnesota State University - Moorhead
United States
marryand@mnstate.edu
Bob Cosmano
WoWiWe Instruction Co LLC
United States
cosmano@wowiwe.net
Abstract: The Virtual Cell is an interactive, 3-dimensional visualization of a bio-environment
designed to teach the structure and process of Cellular Biology. To the students, the Virtual Cell
looks like an enormous navigable space populated with 3D organelles and other structures. They
explore this space using a ‘fly through’ interface, conducting experiments and interpreting them, in
order to complete a sequence of goals. These goals start with identifying organelles, then move on
to more challenging pursuits, including scientific experimentation and question-based assignments
that promote diagnostic reasoning and problem-solving in the authentic visualized context of a 3D
cell body. A software agent ‘guide’ makes assignments, and software tutors assist when the
students require remediation. The instruction is designed to comply with Next Generation Science
Standards, and the game-like aspects are fun and engaging.
Introduction
Student understanding of the structure and function of the cell is essential to modern biology education.
Learning the physical aspects of the cell, how they interact, and how they are regulated has largely replaced the
traditional approaches to teaching biology. A complex, multidimensional environment, time and space are critical
factors that determine cellular events. Communicating this multi-dimensionality in a 2-D medium is difficult.
HS-LS1-c
HS-LS1-h
Organization for Matter and Energy Flow in Organisms
Develop a model to support explanations for how photosynthesis transforms light energy into
stored chemical energy.
HS-LS1-i
Addresses how carbon, hydrogen, and oxygen from sugar molecules produced through
photosynthesis may combine with other elements to form amino acids and other large carbonbased molecules.
HS-LS1-j
Use a model to represent and support the explanation that cellular respiration is a chemical
process whereby the bonds of food molecules and oxygen molecules are broken and the bonds
in new compounds are formed resulting in a net transfer of energy.
HS-LS1-b (*)
Critically read scientific literature and produce scientific writing and/or oral presentations that
communicate how DNA sequences determine the structure and function of proteins, which
carry out most of the work of the cell.
(*) Module based on this standard in development and not presented here.
Table 1: Next Generation Science Standards (NGSS 2013) incorporated in the Virtual Cell
The Virtual Cell is an immersive virtual environment (IVE), where students ‘fly around’ a 3D world and
practice the role of a cell biologist in standards-driven (Tab. 1), goal-oriented surroundings (White, McClean &
Slator 1999). Students learn fundamental concepts of cell biology and strategies for diagnostic problem solving.
This pedagogical approach provides students with authentic experiences that include elements of practical
experimental design and decision making, working individually, or with others. Large scale classroom-based
experimental studies have shown significant learning gains with IVEs (McClean, et al. 2001; Slator, et al. 2005).
Background
An educational game should both engage and inform. Players should learn concepts and skills through
playing the game, and this learning should be transferable to contexts outside the game. The challenge is to build a
game of sufficiently interesting complexity while remaining true to the subject matter. A simulated environment
must react appropriately to all player actions. Without this consistency, the game will fail the ultimate test: students
will not play it (Slator & Chaput 1996).
The World Wide Web Instructional Committee (WWWIC) at North Dakota State University (NDSU) is an
ad hoc group doing research aimed at developing these engaging educational games: immersive virtual
environments (IVEs) designed to study learning in simulated worlds (Slator et al. 2006). WWWIC IVE systems
share a common signature on design and implementation levels (Borchert et al. 2010). Pedagogical design features
qualities such as: immersive, spatially oriented, game-like, highly interactive, exploratory, goal-oriented, learn by
doing, and role-based. These principles are echoed by others in the field (Gaydos & Squire 2010).
Virtual role-playing environments are powerful mechanisms of instruction, when learning how to play and
win the game transfers to real-world concepts and procedures (Bransford & Schwartz 1999; Schönborn & Bögeholz
2009). For over ten years, WWWIC at NDSU has developed environments to enhance student understanding of
geology (Planet Oit), environmental science (eGEO), cellular biology (Virtual Cell), programming languages
(ProgrammingLand), micro-economics (Dollar Bay), Anthropology (On-A-Slant), and history (Blackwood). These
complex systems present a number of opportunities and challenges, with players afforded a role-based, multi-user,
‘learn-by-doing’ experience, and software agents acting as both environmental effects and tutors. Additional
potential arises amid the possibilities of multi-user cooperation and collaboration.
To further WWWIC’s goals, a small business was established to commercialize the efforts through the
Small Business Innovation and Research grant program. WoWiWe Instruction Co. (http://www.wowiwe.net) was
founded in 2002 as an educational media company devoted to immersive virtual environments for education,
authentic instruction, scenario-based assessment, and changing education in our schools. Its primary mission is to
provide experimentally validated educational experiences to parents, students, teachers, and members of the general
public.
The Virtual Cell Animation Project
In the early stages of the Virtual Cell project at NDSU, illustrative 3D animations of the biological
processes were prototyped in VRML using software like Cosmo Worlds in an iterative process that employed
content matter experts in conjunction with graphic artists and 3D modeling professionals. Working closely together,
an accurate and authentic model would be produced over the course of weeks. Once the 3D modeling was complete,
the component elements were exported and integrated into an equivalent simulation, developed to operate on a
server. Client software was developed to access the simulation on the networked server machine, and a software
environment was developed on the server to support the simultaneous connections by multiple users. Thus, from the
earliest days, the Virtual Cell has been a Multi-player Online Game.
To reiterate, the process was to first build a 3D model, then export 3D material to an online simulation. As
time progressed, the project gained access to better animation software (Autodesk, formerly Alias Wavefront,
Maya). The leap in quality, from VRML editors to Maya, had the additional benefit of providing an animation
rendering component, and it was immediately observed that short animations could be produced in Maya by simply
capturing a ‘fly through’ of the 3D model. This prompted a spin-off effort. While the Virtual Cell project has
continued to work on producing interactive simulations, the Virtual Cell Animation Project has begun producing a
series of non-interactive movie style videos that are available on YouTube and at http://vcell.ndsu.edu/animations/.
Implementation
The Virtual Cell provides an authentic problem-solving experience, engaging students in actively learning
structures and functions of eukaryotic cells. The Virtual Cell simulation implements a series of cellular
environments with a variety of assays, molecules, proteins, and tools providing an experimental context for the
student. A virtual avatar acts as a guide and gives out assignments, and intelligent software tutor agents provide
content and problem solving advice (Slator 1999). The Virtual Cell supports multi-user collaborations, where both
students and teachers from remote sites can communicate with each other and work together on shared goals.
The Virtual Cell consists of three software elements: 1) a collection of 3D worlds representing the interior
of a variety of virtual cells and the interior of cellular organelles; 2) a server and database that contains the textual
material (help file and experimental output data), and controls the single and multi-user interactivity; and 3) a client
interface for students to interact with the system from school or home. The 3D worlds provide the visual context
while the server, database, and interface control the interactions. The simulation operates on a network server that
permits multiple users to simultaneously join the 3D worlds and their associated data.
The client user interface has support for both the Mac and PC platforms using the Unity Technologies 3D
game engine. This interface communicates with the server and consists of two primary modes. The main mode is a
fully interactive 3D world operated through a submarine cockpit. In this mode, students interact with cell structures,
observe process simulations, run experiments, collect results, and meet other players flying in their submarines (Fig.
1). They get immediate access to how much ATP fuel their submarine has left, their current goal, what kinds of
torpedoes they can shoot, the name of the cell they are in and its current health, and a reticle (cross-hairs) to aim
their attention. Second, a menu mode contains supplementary information about the game. There are interfaces for
player inventory, goals, game preferences, an encyclopedia of important terms, concepts and procedures, and a chat
interface to type messages to other connected players.
The Virtual Cell consists of a series of fully explorable cell bodies and sub-cellular structures. The initial
point of entry is at the cockpit of a miniature submarine sitting in a needle waiting for injection into a plant cell.
Here the student learns the basic controls and user interface elements of the game and encounters their first scientific
mentor (a software agent ‘guide’) represented as a robot, and receives a specific assignment. From there, the student
visits the interior of the first “identification” virtual cell, where they perform simple experiments (assays), and learn
the basic physical and chemical features of the cell and its components. As the student progresses through the levels
of the module, the robot gives them more assignments and provides feedback on their progress.
Organelle Identification Module
While in the submarine, the learners receive specific assignments from robot tutors. This is a hallmark of
these systems – learners are always in possession of a goal, and there is never any doubt about what goal they are
supposed to be working on. To achieve these goals, students perform the experiments necessary to learn the physical
and chemical features of the cell and its components. In the submarine, learners are supplied with a toolbox of
measuring devices that assay various cellular processes. These tools include such things as protein assays,
molecular substrates, proteins meant to repair broken complexes, and enzyme assays.
The first module of the Virtual Cell is an ‘identification game’ populated with sub-cellular components:
nucleus, endoplasmic reticulum, golgi apparatus, mitochondrion, chloroplast, ribosome, and vacuole. The user
‘flies’ among these organelles and uses virtual instruments to conduct experiments designed to identify them. For
example, the learners may confront a nucleus and perform several simple experiments. When performing a glycosol
transferase assay, it shows low levels. When performing a DNA synthesis assay, it shows high levels. The student
may also perform a number of other assays. If the student reports the organelle as a golgi apparatus, the tutor
appears and suggests where to look for information about the DNA synthesis test along with probing questions
designed to encourage the student to do more research before reporting again. Upon correctly identifying the
nucleus, the user is prompted to identify more organelles, until all of them are identified, at which point they can
move on to the Electron Transport Chain or Photosynthesis modules.
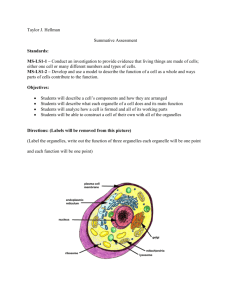
Figure 1: the Virtual Cell’s ID module. The goal is to correctly identify seven organelles inside an
archetypical plant cell. Here, the player is aiming the DNA Synthesis assay at an unidentified
organelle in an attempt to find the nucleus. Another player hovers nearby in their submarine.
Advanced Modules
As learners progress through the modules they are also able to travel into linked 3D worlds that represent
the interior of selected cellular organelles. Further experimentation inside each organelle allows the students to
learn about its specific functions (Wu 1998). More advanced modules of the Virtual Cell are designed for the study
of the electron transport chain within the mitochondria and photosynthesis within the chloroplast, as well as others,
such as the gene expression functions of transcription and translation. The approach taken in each case is the same.
Learners begin by entering a healthy organelle, such as a mitochondrion that is producing ATP visibly into
the 3D world. The student is able to view a simulation of the process including the complexes, reactions, and
movement of molecules in a 3D environment. All complexes are interactive and contain descriptions from the
embedded encyclopedia describing what they do. Students can fire substrate torpedoes at the chains, resulting in the
restart of the simulation.
Upon leaving the healthy mitochondrion, they are tasked with repairing a broken one. As before, upon
entering the mitochondrion, the student can observe the electron transport chain process. However, the process does
not complete as before. Inserting a substrate into the chain causes the process to advance to the next step, but what
substrates to use? They may learn from a lecture, a textbook, the encyclopedia or a tutoring agent. One of the
complexes will not accept the appropriate substrate, indicating that it is broken. Students then gather the necessary
replacement and restart the electron transport chain, healing the cell and completing their task.
Mini-games
Good lesson plans contain contingencies for situations where students finish early or for those who have
trouble understanding lesson concepts. To address this, WoWiWe develops mini-games to go along with each lesson
module. These mini-games are meant to reinforce the basic concepts of a module and provide motivation to practice
particular skills without necessarily using the same submarine-flying-around modality as the original lesson. For
example, the Virtual Cell’s module on the electron transport chain (ETC) has a mini-game that is an orientation
puzzle, where the objective is to rebuild one or more ETCs in a mitochondrion by moving and aligning the
complexes. When enough chains are functioning properly, ATP production is steady enough to win the level and
advance to the next.
Learning and Assessment
IVEs such as the Virtual Cell are intended to provide rich and interesting spaces for exploration and
discovery. Student activities are partitioned into modules that are organized around a set of goals, which are
accomplished by completing an accompanying set of tasks. Tasks are typically activities that require application of a
skill to complete. Completing tasks and achieving goals are the means by which a student progresses through the
simulation. They are also a measurable means for demonstrating the mastery of a skill set.
This method of measuring student progress is often called “embedded assessment”. Students are assigned
goals whose nature and sequence are designed by content matter experts to be appropriate to their experience and
level in the game. Goals are assigned point values, and players accumulate objectively measured scores as they
achieve their goals. The goals are taken from a principled set, where easier goals are followed by more advanced
ones. This is formative assessment as the simulation proceeds. Students demonstrate learning by achieving goals.
The goals are chosen to align with national science education standards (NGSS 2013).
Summative assessment is provided at the end of each game module. These take the form of “mission
reports” where players fill out forms detailing their accomplishments in the module. Filling out each form entails
answering questions about the biological structures and processes found in each module. These reports currently
take the form of multiple choice exams at the conclusion of the Electron Transport Chain and Photosynthesis
modules.
Conclusion
The over-arching pedagogical goal of this project is to support role-based learning (Slator et al. 2001)
where students are presented with authentic problems that promote their ability to think and act like a biologist.
This means developing a curriculum of authentic tasks and goals and providing an inventory of authentic tools and
instruments to achieve these goals. In addition, the aim is to provide these experiences at a distance, using an
Internet delivery mechanism. Further, to provide a self-paced element, there is a need to provide extensive online
help and agent-based software tutors to supplement human intervention and remediation. Finally, it is extremely
desirable in modern pedagogy to provide opportunities for group interaction and collaborative problem-solving. In
order to support these pedagogical goals, it is necessary to develop a highly interactive and dynamic simulation of
cellular processes, hosted on the Internet, supporting multiple users.
WWWIC and WoWiWe Instruction Co continue to develop the Virtual Cell. A Photosynthesis module is
near completion and planned future modules include Transcription, Translation, and Cell Division. Each module
will be developed with an eye towards a unique, fun, and experimentally validated approach towards learning. The
Virtual Cell can be visited at http://www.wowiwe.net
References
Borchert, O., Brandt, L., Hokanson, L., Slator, B. M., Vender, B., & Gutierrez, E.J. (2010). Principles and Signatures in Serious
Games for Science Education, in Gaming and Cognition: Theories and Practice from the Learning Sciences Edited by: Richard
Van Eck. IGI Global. pp. 312-338.
Bransford, J. D., & Schwartz, D. L. (1999). Chapter 3: Rethinking Transfer: A Simple Proposal with Multiple Implications.
Review of Research in Education. 24 (1). 61-100.
Gaydos, M., & Squire, K. (2010). Citizen Science : Designing a Game for the 21st Century. In R. Van Eck
(Ed.), Interdisciplinary Models and Tools for Serious Games: Emerging Concepts and Future Directions (pp. 289-305). Hershey,
PA: Information Science Reference. doi:10.4018/978-1-61520-719-0.ch012
McClean, P., Saini-Eidukat, B., Schwert, D., Slator, B., & White, A. (2001). Virtual Worlds in Large Enrollment Biology and
Geology Classes Significantly Improve Authentic Learning. In: Selected Papers from the 12th International Conference on
College Teaching and Learning (ICCTL-01), Jack A. Chambers, Editor). Jacksonville, FL: Center for the Advancement of
Teaching and Learning. April 17-21, pp. 111-118.
NGSS (2013). The Next Generation Science Standards. Retrieved January 25, 2013 from http://www.nextgenscience.org/nextgeneration-science-standards
Schönborn, K. J., & Bögeholz, S. (2009). Knowledge Transfer in Biology and Translation across External Representations:
Experts' Views and Challenges for Learning. International Journal of Science and Mathematics Education. 7: 931-955.
Slator, B. M., & Chaput, H. (1996). Learning by Learning Roles: A Virtual Role-Playing Environment for Tutoring. Proceedings
of the Third International Conference on Intelligent Tutoring Systems (ITS'96). Montréal: Springer-Verlag, June 12-14, pp. 668676. (Lecture Notes in Computer Science, edited by C. Frasson, G. Gauthier, A. Lesgold).
Slator, B. M. (1999). Intelligent Tutors in Virtual Worlds. Proceedings of the 8th International Conference on Intelligent
Systems. Denver, CO. June 24-26, pp. 124-127.
Slator, B. M., Clark, J., Juell, P., McClean, P., Saini-Eidukat, B., Schwert, D. P., & White A. R. (2001). Research on Role-based
Learning Technologies. Proceedings of the First IEEE International Conference on Advanced Learning Technologies (ICALT01). Madison, WI, Aug. 6-8. pp. 37-41
Slator, B., Dong, A., Erickson, K., Flaskerud, D., Halvorson, J., Myronovych, O., McClean, P., Saini-Eidukat, B., Schwert, D. P.,
White, A., & Terpstra. J. (2005). Comparing Two Immersive Virtual Environments for Education. Proceedings of E-Learn 2005,
Vancouver, Canada, October 24-28.
Slator, B. M., Beckwith, R., Brandt, L., Chaput, H., Clark, J. T., Daniels, L. M., Hill, C., McClean, P., Opgrande, J., SainiEidukat, B., Schwert, D. P., Vender, B., & White, A. R. (2006). Electric Worlds in the Classroom: Teaching and Learning with
Role-Based Computer Games. New York: Teachers College Press. Columbia University. 192 pages.
White, A. R., McClean, P. E., & Slator, B. M. (1999). The Virtual Cell: An Interactive, Virtual Environment for Cell Biology.
World Conference on Educational Media, Hypermedia and Telecommunications (ED-MEDIA 99), June 19-24, Seattle, WA.
Wu, Y. (1998). Simulation of the biological process of steroid hormones with VRML. Master’s Thesis. North Dakota State
University, Fargo.
Acknowledgements
This research was supported in part by an SBIR grant from the National Institutes of Health, National Center for Research
Resources (NIH-2R44RR024779-02A1). Artists responsible for interface design, production graphics, and screenshots in this
paper are Katy Cox (kcox@wowiwe.net) and Andre Pilch (apilch@wowiwe.net).