Chapter 7: Gene Expression: The Flow of Genetic Information from
advertisement

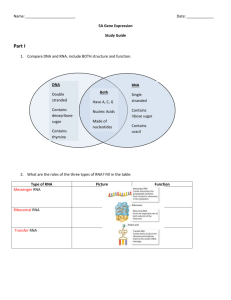
Chapter 7: Gene Expression: The Flow of Genetic Information from DNA via RNA to Protein Outline I. The genetic code: how precise groupings of the 4 nucleotides specify 20 amino aAcids A. In the genetic code, a triplet codon represents each amino acid B. Mapping studies confirmed that a gene’s nucleotide sequence is colinear with a polypeptide’s amino acid sequence C. Genetic analysis revealed that nonoverlapping codons are set in a reading frame 1. A codon is composed of more than one nucleotide 2. Each nucleotide is part of only a single codon 3. A codon is composed of three nucleotides, and the designated starting point for each gene establishes the reading frame for these triplets 4. Most amino acids are specified by more than one codon, which makes the genetic code a “degenerate” code D. Cracking the code: biochemical manipulations revealed which codons represent which amino acids 1. The discovery of messenger RNAs, molecules for transporting genetic information 2. Synthetic mRNAs and in vitro translational systems made it possible to discover which codons designate which amino acids 3. The 5’-to-3’ direction in mRNA corresponds to the N-terminal-to-C- terminal direction in a polypeptide 4. Nonsense codons cause termination of a polypeptide chain E. The genetic code: a summary F. Using genetics to verify the code G. The genetic code is almost, but not quite, universal II. Transcription: RNA polymerase synthesizes an mRNA copy of a gene’s template strand A. Details of the process B. In eukaryotes, mRNA processing after transcription produces a mature messenger RNA 1. Adding a methylated cap at the 5’ end and a poly-A tail at the 3’ end 2. RNA splicing removes sequences known as introns from the primary transcript 3. Alternative splicing often produces different mRNAs from the same primary transcript III. Translation: base-pairing between mRNA and tRNAs directs assembly of a polypeptide on the ribosome A. Transfer RNAs mediate the translation of mRNA codons to amino acids 1. tRNA structure: A compact “L” carrying an anticodon at one end and an amino acid at the other 2. Base-pairing between an mRNA codon and a tRNA anticodon directs incorporation of an amino acid into a growing polypeptide 3. Wobble: Some tRNAs can recognize more than one codon B. Ribosomes are the sites of polypeptide synthesi 1. Ribosomes are complex structures composed of RNA and protein 2. Different parts of the ribosome have different functions C. The mechanism of translation; there are some significant differences in translation between prokaryotes and eukaryotes D. Processing after translation can change a polypeptide’s structure IV. Comprehensive example: a computerized analysis of gene expression in C. elegans V. How mutations affect gene expression A. Mutations in a gene’s coding sequence can alter the gene product 1. Silent mutations do not alter the amino acid specified 2. Missense mutations replace one amino acid with another 3. Nonsense mutations change an amino-acid-specifying codon to a stop codon 4. Frameshift mutations result from the insertion or deletion of nucleotides within the coding sequence B. Mutations in a gene outside the coding sequence can also alter gene expression C. Mutations in genes encoding the molecules that implement expression may affect transcription, mRNA splicing, or translation 1. Mutations altering genes encoding proteins or RNAs involved in gene expression are usually lethal 2. Mutations in tRNA genes can suppress mutations in protein-coding genes Essential Concepts Gene expression is the process by which cells convert the DNA sequence of a gene to the RNA sequence of a transcript, and then decode the RNA sequence to the amino acid sequence of a polypeptide. The nearly universal genetic code consists of 64 codons composed of 3 nucleotides apiece. 61 of these codons specify amino acids, while the remaining 3 UAA, UAG, and UGA are nonsense or stop codons that do not specify an amino acid. a. The code is degenerate: More than one codon specifies every amino acid except methionine and tryptophan. b. AUG in the context of a ribosome binding site is the initiation codon; it establishes a reading frame that determines the grouping of nucleotides into triplet codons. c. The code is nonoverlapping. Within a reading frame, the first three nucleotides constitute one codon, the next three, the second codon, and so forth. Gene expression based on the genetic code produces the colinearity of a gene’s nucleotide sequence and a protein’s sequence of amino acids. Transcription is the first stage of gene expression. During transcription, RNA polymerase synthesizes a single stranded mRNA transcript from a DNA template. a. RNA polymerase initiates transcription by binding to the promoter sequence of the DNA and unwinding the double helix to expose bases for pairing. b. RNA polymerase extends the mRNA in the 5’-to-3’ direction by catalyzing formation of phosphodiester bonds between successively aligned nucleotides. c. Terminator sequences in the RNA cause RNA polymerase to dissociate from the DNA. d. In prokaryotes, the primary transcript is the mRNA that guides polypeptide synthesis. In eukaryotes, RNA processing after transcription produces a mature mRNA that travels from the nucleus to the cytoplasm to direct polypeptide synthesis. a. RNA processing adds a methylated cap to the 5’ end and a poly-A tail to the 3’ end of the eukaryotic mRNA. b. The spliceosome removes introns from the primary transcript and precisely splices together the remaining exons. Alternative splicing makes it possible to produce different mRNAs from the same primary transcript. Translation is the stage of gene expression when the cell synthesizes proteins according to instructions in the mRNA. a. tRNAs carry amino acids to the translation machinery. Aminoacyl tRNA synthetases connect amino acids to their corresponding tRNAs. Each tRNA molecule has an anticodon complementary to the mRNA codon specifying the amino acid it carries. Because of wobble, some tRNA anticodons recognize more than one mRNA codon. b. Translation occurs on complex molecular machines called ribosomes. Ribosomes have two binding sites for tRNAs ð P and A, and an enzyme known as peptidyl transferase that catalyzes formation of a peptide bond between amino acids carried by the tRNAs at these two sites. c. Initiation: To start translation, part of the ribosome binds to a ribosome binding site on the mRNA, which includes the AUG initiation codon. Special initiating tRNAs with codons complementary to AUG carry the amino acid f-met in prokaryotes or met in eukaryotes to the ribosomal P site. This amino acid will become the N-terminus of the growing polypeptide. d. Elongation: When the carboxyl group of the amino acid connected to a tRNA at the ribosome’s P site becomes attached through a peptide bond to the amino acid carried by the tRNA at the A site, the ribosome travels three nucleotides toward the 3’ end of the mRNA. This movement exposes the next codon and allows the next round of amino acid addition. This mechanism of translation dictates that the 5’-3’ direction in the mRNA corresponds to the N-terminus-to-C-terminus direction in the polypeptide under construction. e. Termination: When the ribosome encounters in-frame nonsense codons, it ends translation by releasing the mRNA and disconnecting the complete polypeptide from the tRNA. Processing after translation may alter a polypeptide by adding or removing chemical constituents to or from particular amino acids, or by cleaving the polypeptide into smaller molecules. Mutations affect gene expression in several ways. a. Mutations in a gene may modify the message encoded in a sequence of nucleotides. Silent mutations usually change the third letter of a codon and have no effect on polypeptide production. Missense mutations change the codon for one amino acid to the codon for another amino acid and thereby direct incorporation of a different amino acid. Nonsense mutations change a codon for an amino acid to a stop codon, causing synthesis of a truncated polypeptide. Frameshift mutations change the reading frame of a gene, altering the identity of amino acids downstream of the mutation. b. Mutations outside of coding sequences that alter signals required for transcription, mRNA splicing, or translation, will also disrupt gene expression. c. Mutations in genes encoding molecules of the gene expression machinery are often lethal. Exceptions arise when another gene supplies a molecule with a similar function. Among these exceptions are mutations in tRNA genes that suppress mutations in polypeptide-encoding genes.