Word file (51.5 KB )
advertisement

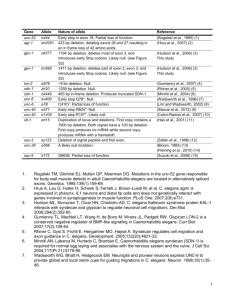
Supplementary table I Plasmid cDNA library pACT-2 pAS2-1 pGEX-4T pLP132 pLP185 L4440 (feeding vector) pLP119 (pBKS) pAS2-1 pACT-2 pAS2-1 pACT-2 pAS2-1 pACT-2 pAS2-1 pACT-2 pAS2-1 pACT-2 pAS2-1 pACT-2 pAS2-1 pSG15 pLP168 (pBKS) pLP170 (pAS2-1) pLP173 pLP36 (feeding) Insert GST GST-MEI-1 GST-RBX-1 CUL-3 CUL-2 MEL-26 MEL-26 MEI-2 MEI-2 mel-26(ct61sb4) mel-26(ct61sb4) mel-26(sb45) mel-26(sb45) MEI-1 MEI-1 mei-1(ct46gf) mei-1(ct46gf) GST-MEL-26 CUL-3S50AF51A CUL-3S50AF51A cul-3 N-term CUL-3 Source /Ref Invitrogen Clontech Clontech Pharmacia This study This study 1 This study E. Kipreos This study This study This study This study This study This study This study This study This study This study This study This study This study This study This study This study 2 -1- Supplementary Figure 1: Examination of the microtubule network by indirect immunofluorescence in a or543ts mutant embryo as compared to wild-type. Confocal micrograph of -tubulin and DNA in a fixed one cell embryo. Note that in mel-26(or543ts) embryos many astral microtubules do not reach the cortex. Supplementary Figure 2: (A) MEI-1 interacts with itself, as expected since the corresponding katanin subunits form hexameric rings, as well as with the product of the gain-of-function allele mei-1(ct46gf). These data are consistent with previous observations that MEI-1(gf) results in ectopic mitotic expression of both MEI-1(+) 3and MEI-2 4. We also examined interactions of the MEL-26 mutant sb45 and ct61sb4 5. While MEL-26(+) shows self interactions, the truncation mutant ct61sb4, which behaves as a genetic null at elevated temperatures, did not interact with MEL26(+), MEI-1 or MEI-1(gf). However, the dominant-negative mel-26 allele sb45, although not able to interact with MEI-1(+), still binds MEL-26(+). This is consistent with the dominantnegative properties of sb45: binding to MEL-26(+) likely results in a complex that cannot bind MEI-1(+).The interactions were determined on yeast colonies using the -galactosidase filter assay. (B) The expression of wild-type CUL-3 and the CUL-3LAAE mutants fused to the DNAbinding domain of Gal4 was tested by westernblot using anti-CUL-3 antibodies. Supplementary Figure 3: A single BTB-domain-containing polypeptide may bridge some cullins to their ubiquitination substrates. The topologies of the Skp1/F-box and ElonginC/SOCS-box heterodimer interfaces suggest that a two domain single polypeptide with a four helix inter-domain linker (top row) may have been the evolutionary precursor of both classes of heterodimer. Separation between the second and third helices or between the first and second helices could have lead to the Skp1/F-box and ElonginC/SOCS-box families respectively (second and third rows, with example proteins indicated). The relevant structural similarities are demonstrated by the crystal structures of the Skp1/Cdc4 and Skp1/Skp2 structures (second row; 6 7) and the ElonginC/VHL structure (fourth row; 8), for which numbered helices are shown. The BTB domain/cullin interactions indicated are discussed in the text and elsewhere 9. Such evolutionary speculation strengthens the hypothesis that an extant single polypeptide, such as MEL-26, which contains both a BTB domain and another protein-protein interaction domain may function as an adapter between a cullin, CUL-3, and its substrate, MEI-1 (fifth row). Other BTB domain-containing proteins may have a similar function, with one possible example shown, mayven 10 (sixth row). -2- Supplementary references 1. Timmons, L. & Fire, A. Specific interference by ingested dsRNA. Nature 395, 854 (1998). 2. Pintard, L. et al. Neddylation and deneddylation of CUL-3 is required to target MEI1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr. Biol. 13, 911–921 (2003). 3. Clandinin, T. R. & Mains, P. E. Genetic studies of mei-1 gene activity during the transition from meiosis to mitosis in Caenorhabditis elegans. Genetics 134, 199–210 (1993). 4. Srayko, M., Buster, D. W., Bazirgan, O. A., McNally, F. J. & Mains, P. E. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 14, 1072–1084 (2000). 5. Dow, M. R. & Mains, P. E. Genetic and molecular characterization of the Caenorhabditis elegans gene, mel-26, a postmeiotic negative regulator of mei-1, a meiotic- specific spindle component. Genetics 150, 119–128 (1998). 6. Orlicky, S., Tang, X., Willems, A., Tyers, M. & Sicheri, F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112, 243–256 (2003). 7. Schulman, B. A. et al. Insights into SCF ubiquitin ligases from the structure of the Skp1Skp2 complex. Nature 408, 381–386 (2000). 8. Stebbins, C. E., Kaelin, W. G., Jr & Pavletich, N. P. Structure of the VHL-ElonginCElonginB complex: implications for VHL tumor suppressor function. Science 284, 455– 461 (1999). 9. Kamura, T. et al. Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 276, 29748–29753 (2001). 10. Soltysik-Espanola, M. et al. Characterization of Mayven, a novel actin-binding protein predominantly expressed in brain. Mol. Biol. Cell 10, 2361–2375 (1999). -3-