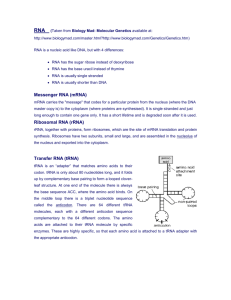
RNA Categories:
mRNA - Messenger RNA, hnRNA, heterogeneous nuclear RNA, ribonucleoproteins,
RNPs, interference, transcription
tRNA - Transfer RNA, translation, decoding, acceptor stem
snRNA - Small nuclear RNA, nucleus, small nuclear ribonucleoproteins, (SNRNP),
snurps.
rRNA - Ribosomal RNA, 18 s, 5.8 s, 28 s, 5 s
Preprocessing phase
Collected Abstracts
Source: Pubmed
Keyword search: “Messenger RNA”
1: Mol Reprod Dev. 2005 Dec;72(4):471-82.
Nuclear lamin antigen and messenger RNA expression in bovine in vitro
produced and nuclear transfer embryos.
Hall VJ, Cooney MA, Shanahan P, Tecirlioglu RT, Ruddock NT, French AJ.
Centre for Early Human Development, Monash Institute of Medical
Research, Monash University, Clayton, Victoria, Australia.
The nuclear lamina is a complex meshwork of nuclear lamin filaments
that lies on the interface of the nuclear envelope and chromatin and is
important for cell maintenance, nucleoskeleton support, chromatin
remodeling, and protein recruitment to the inner nucleolus. Protein and
mRNA patterns for the major nuclear lamins were investigated in bovine
in vitro fertilized (IVF) and nuclear transfer embryos. Expression of
lamins A/C and B were examined in IVF bovine germinal vesicle (GV)
oocytes, metaphase II oocytes, zygotes, 2-cell, 8-cell, 16-32-cell
embryos, morulae, and blastocysts (n = 10). Lamin A/C was detected in
9/10 immature oocytes, 10/10 zygotes, 8/10 2-cell embryos, 4/10 orulae,
10/10 blastocysts but absent during the maternal embryonic transition.
Lamin B was ubiquitously expressed during IVF preimplantation
development but was only detected in 4/10 GV oocytes. Messenger RNA
expression confirms that the major lamins, A/C and B1 are expressed
throughout preimplantation development and transcribed by the embryo
proper. Lamin A/C and B expression were observed (15 min, 30 min, 60
min, 120 min) following somatic cell nuclear transfer using adult
fibroblasts and at the 2-cell, 8-cell, 16-32-cell, morula and
blastocyst stage (n = 5). Altered expression levels and localization of
nuclear lamins A/C and B was determined in nuclear transfer embryos
during the first 2 hr post fusion, coincidental with only partial
nuclear envelope breakdown as well as during the initial cleavage
divisions, but was restored by the morula stage.
This mechanical and molecular disruption of the nuclear lamina provides
key evidence for incomplete nuclear remodeling and reprogramming
following somatic cell nuclear transfer. Mol. Reprod. Dev. (c) 2005
Wiley-Liss, Inc.
PMID: 16161164 [PubMed - in process]
2: Am J Surg. 2005 Nov;190(5):739-45.
Effect of cyclophilin A on gene expression in human pancreatic cancer
cells.
Li M, Wang H, Li F, Fisher WE, Chen C, Yao Q.
Molecular Surgeon Research Center, Baylor College of Medicine, Houston,
TX, USA; Elkins Pancreas Center, Michael E. DeBakey Department of
Surgery, Baylor College of Medicine, One Baylor Plaza, Mail stop: NAB2010, Houston, TX 77030, USA.
BACKGROUND: We previously found that cyclophilin A (CypA) is
overexpressed in human pancreatic cancer cells and stimulates cell
proliferation through CD147. In this study, we further investigated the
effect of CypA on gene expression of several key molecules that are
involved in pancreatic cancer cell proliferation. METHODS: Human
pancreatic cancer cell lines (Panc-1, MIA PaCa-2, and BxPC-3) and
human pancreatic ductal epithelial (HPDE) cells were used. The
messenger RNA (mRNA) levels of CypA, CypB, CD147, neuropilins (NRPs),
vascular endothelial growth factor (VEGF), and VEGF receptors upon the
treatment of exogenous recombinant human CypA were determined by realtime reverse-transcription polymerase chain reaction. RESULTS:
Exogenous human recombinant CypA reduced the mRNA levels of NRP-1 and
VEGF, but not endogenous CypA, CypB, and CD147, in Panc-1, MIA PaCa-2,
and BxPC-3 cells. In contrast, HPDE cells showed a decrease of
endogenous CypA and CD147 mRNA, but not detectable changes of CypB,
NRPs, and VEGF mRNA levels upon exogenous CypA treatment. CONCLUSIONS:
These data show that exogenous CypA downregulates NRP-1 and VEGF
expression in pancreatic cancer cells. This effect is different in
normal HPDE cells. Thus, soluble CypA may affect cell growth of
pancreatic cancer.
PMID: 16226951 [PubMed - in process]
3: Biomaterials. 2005 Nov;26(33):6643-51.
A silanized hydroxypropyl methylcellulose hydrogel for the threedimensional culture of chondrocytes.
Vinatier C, Magne D, Weiss P, Trojani C, Rochet N, Carle GF, VignesColombeix C, Chadjichristos C, Galera P, Daculsi G, Guicheux J.
INSERM EM 9903, Research Center on Materials with Biological Interest,
School of Dental Surgery, University of Nantes, 1-Place Alexis
Ricordeau 44042, Nantes Cedex 1, France.
Articular cartilage has limited intrinsic repair capacity. In order to
promote cartilage repair, the amplification and transfer of autologous
chondrocytes using three-dimensional scaffolds have been proposed. We
have developed an injectable and self-setting hydrogel consisting of
hydroxypropyl methylcellulose grafted with silanol groups (Si-HPMC).
The aim of the present work is to assess both the in vitro
cytocompatibility of this hydrogel and its ability to maintain
a chondrocyte-specific phenotype. Primary chondrocytes isolated from
rabbit articular cartilage (RAC) and two human chondrocytic cell lines
(SW1353 and C28/I2) were cultured into the hydrogel. Methyl tetrazolium
salt (MTS) assay and cell counting indicated that Si-HPMC hydrogel did
not affect respectively chondrocyte viability and proliferation.
Fluorescent microscopic observations of RAC and C28/I2 chondrocytes
double-labeled with cell tracker green and ethidium homodimer-1
revealed that chondrocytes proliferated within Si-HPMC. Phenotypic
analysis (RT-PCR and Alcian blue staining) indicates that chondrocytes,
when three-dimensionnally cultured within Si-HPMC, expressed
transcripts encoding type II collagen and aggrecan and produced
sulfated glycosaminoglycans. These results show that Si-HPMC allows the
growth of differentiated chondrocytes. Si-HPMC therefore appears as a
potential scaffold for three-dimensional amplification and transfer of
chondrocytes in cartilage tissue engineering.
PMID: 15950277 [PubMed - indexed for MEDLINE]
4: Clin Chim Acta. 2005 Nov;361(1-2):182-90.
Messenger RNA expression of glomerular podocyte markers in the urinary
sediment of acquired proteinuric diseases.
Szeto CC, Lai KB, Chow KM, Szeto CY, Yip TW, Woo KS, Li PK, Lai FM.
Department of Medicine and Therapeutics, Prince of Wales Hospital, The
Chinese University of Hong Kong, Shatin, Hong Kong, China.
BACKGROUND: Podocyte slit diaphragm plays an important role in the
control of glomerular permeability. We hypothesize that studying the
gene expression profile of podocyte in urinary sediment may provide
diagnostic and prognostic information on acquired proteinuric diseases.
METHODS: We studied 28 patients who required kidney biopsy for acquired
proteinuric diseases (diabetic glomerulosclerosis, 9 cases; IgA
nephropathy, 10 cases; minimal change disease, 5 cases; membranous
nephropathy, 5 cases). We also studied 10 cases of diabetic
microalbuminuria and 9 healthy controls. The mRNA expressions of
nephrin (NephRNA), podocin (PodRNA) and synaptopodin (SynRNA) in
urinary sediment were measured by real time quantitative PCR. After
recruitment, all patients were followed for at least 12 months.
RESULTS: There were significant differences in the NephRNA and PodRNA
in the urinary sediment between diagnosis groups (p<0.005). On the
other hand, SynRNA was only marginally significant between diagnosis
groups (p<0.05). Although statistically significant, the degree of
proteinuria had only modest correlations with the urinary expression of
nephrin. After a median follow up for 23 months, there was a
significant correlation between the rate of decline in renal function
and NephRNA (r=0.559, p=0.001) and PodRNA (r=0.530, p=0.002), but not
SynRNA (r=0.054, p=NS). The correlation remained statistically
significant after multivariate analysis to adjust for the degree of
proteinuria and initial renal function. CONCLUSIONS: Urinary mRNA
expression of podocyte markers, such as nephrin and podocin, are
significantly different between proteinuric disease categories.
Further, NephRNA and PodRNA correlated with the rate of decline in
renal function. Our results suggest that urinary podocyte gene
expression may be a useful non-invasive tool which provides additional
information for the management of proteinuric diseases.
PMID: 15996647 [PubMed - in process]
5: J Infect Dis. 2005 Nov 1;192(9):1628-33. Epub 2005 Sep 23.
Proinflammatory Responses in the Murine Brain after Intranasal Delivery
of Cholera Toxin: Implications for the Use of AB Toxins as Adjuvants in
Intranasal Vaccines.
Armstrong ME, Lavelle EC, Loscher CE, Lynch MA, Mills KH.
Immune Regulation Research Group, School of Biochemistry and
Immunology, Trinity College, Dublin, Ireland.
Intranasal delivery of vaccines provides an attractive alternative to
parenteral delivery, but it requires appropriate mucosal adjuvants.
Cholera toxin (CT) is a powerful mucosal adjuvant, but it can undergo
retrograde transport to the brain via the olfactory system after
intranasal delivery. We demonstrate that intranasal delivery of CT
increases the expression of interleukin-1 beta , cyclooxygenase-2, and
chemokine messenger RNA in the murine hypothalamus, whereas
parenterally delivered CT has little effect. Our findings suggest that
CT can induce proinflammatory mediators in the brain when it is
administered intranasally but not parenterally, and they raise concerns
about the use of AB toxins as adjuvants in intranasal vaccines.
PMID: 16206078 [PubMed - in process]
6: Int J Cancer. 2005 Oct 20;117(1):82-9.
Subtype-specific expression and genetic alterations of the
chemokinereceptor gene CXCR4 in medulloblastomas.
Schuller U, Koch A, Hartmann W, Garre ML, Goodyer CG, Cama A, Sorensen
N, Wiestler OD, Pietsch T.
Department of Neuropathology, University of Bonn Medical Center, Bonn,
Germany.
Recent findings indicate that the chemokine receptor Cxcr4 is essential
for normal development of the cerebellar cortex. As medulloblastomas
(MBs), the most common malignant brain tumors of childhood, are
believed to arise from neuronal cerebellar precursors, we asked whether
there is a potential role for Cxcr4 in the pathogenesis of MB. RT-PCR
and immunohistochemistry revealed expression of Cxcr4 in different
variants of MBs. Whereas 18/20 classic MBs showed very low levels of
CXCR4 mRNA, high amounts were expressed in 17/18 desmoplastic and 6/7
extensively nodular MBs. In addition, a significant correlation of high
CXCR4 mRNA levels and presence of the neurotrophin receptor p75NTR or
expression of ATOH1 and GLI1 suggests that CXCR4 is a reliable marker
for tumors derived from the cerebellar external granular layer. Because
Cxcr4 is important for migration and cell cycle control of granular
precursors, we screened for mutations in the coding region by SSCP and
gene sequencing. In a series of 90 MBs and 8 MB cell lines, we found
one germline and one somatic mutation resulting in amino acid
substitutions in the first (Ile53Leu) and second (Asp97Asn)
transmembrane regions, respectively. These data suggest that Cxcr4 may
be involved in the pathogenesis of MBs.
Copyright (c) 2005 WileyLiss, Inc. Publication Types:
Multicenter Study
PMID: 15880586 [PubMed - indexed for MEDLINE]
7: Int J Cancer. 2005 Oct 20;117(1):14-20.
Growth-related oncogene produced in human breast cancer cells and
regulated by Syk protein-tyrosine kinase.
Li J, Sidell N.
Division of Research, Department of Gynecology and Obstetrics, Emory
University School of Medicine, Atlanta, GA 30322, USA.
Syk, a nonreceptor type of protein tyrosine kinase widely expressed in
hematopoietic cells, is a candidate suppressor gene in human breast
cancer. Reduced expression of Syk protein is correlated with poor
prognosis, while its overexpression can reduce the malignant phenotype
of breast cancer cells. The mechanism of action of Syk remains unclear.
In this study, we utilized low Syk-expressing, highly invasive MDA-MB231 and high Syk-expressing, less invasive MCF-7 breast cancer cells to
investigate the possibility that part of the functional effects of Syk
are mediated by cytokines known to play roles in cell migration,
invasion or metastasis. Using protein array technology, we determined
that MDA-MB-231 cells secrete a number of cytokines known to regulate
cellular growth and motility. One such cytokine, growth-related
oncogene (GRO), has previously not been described in breast cancer. Of
the compounds detected in the culture supernatant of MDA-MB-231, GRO
was the only one that was significantly altered by modulation of Syk
expression; overexpression of Syk caused a marked reduction in secreted
levels of GRO. Conversely, downregulation of the relatively high levels
of Syk produced in MCF-7 cells upregulated GRO secretion. At the mRNA
level, overexpression of Syk in MDA-MB-231 differentially regulated the
3 GRO isotypes such that message levels of GROalpha and gamma were
downregulated while that of GRObeta was not affected. Matrigel invasion
assays demonstrated a link between Syk expression and resulting GRO
activity in mediating the invasive potential of MDA-MB-231 cells. In
summary, our findings provide evidence that human breast cancer cells
express and secrete GRO and implicate this cytokine as an essential
mediator of the antiinvasive properties of Syk tyrosine kinase.
Copyright (c) 2005 Wiley-Liss, Inc.
PMID: 15880583 [PubMed - indexed for MEDLINE]
8: Int J Cancer. 2005 Oct 20;117(1):51-8.
Periostin is down-regulated in high grade human bladder cancers and
suppresses in vitro cell invasiveness and in vivo metastasis of cancer
cells.
Kim CJ, Yoshioka N, Tambe Y, Kushima R, Okada Y, Inoue H.
Department of Urology, Shiga University of Medical Science, Otsu,
Shiga, Japan.
We have previously reported that expression of periostin mRNA is
markedly reduced in a variety of human cancer cell lines, suggesting
that downregulation of periostin mRNA expression is correlated with the
development of human cancers. In our study, to clarify the role of the
periostin in human bladder carcinogenesis, we examined the expression
of periostin mRNA in normal bladder tissues, bladder cancer tissues and
bladder cancer cell lines by Northern blot analysis and RT-PCR
analysis. Although the expression of periostin mRNA was detected in
100% (5/5) of normal bladder tissues, it was not detected in 3 human
bladder cancer cell lines examined. It was also detected in 81.8%
(9/11) of grade 1, 40.0% (4/10) of grade 2 and 33.3% (4/12) of grade 3
bladder cancer tissues, indicating that downregulation of periostin
mRNA is significantly related to higher grade bladder cancer (p<0.05).
To assess the tumor suppressor function of periostin, we investigated
the ability of periostin gene to suppress malignant phenotypes of a
bladder cancer cell line, SBT31A. Ectopic expression of periostin gene
by a retrovirus vector suppressed in vitro cell invasiveness
of the bladder cancer cells without affecting cell proliferation and
tumor growth in nude mice. Periostin also suppressed in vivo lung
metastasis of the mouse melanoma cell line, B16-F10. Mutational
analysis revealed that the C-terminal region of periostin was
sufficient to suppress cell invasiveness and metastasis of the cancer
cells. Periostin may play a role as a suppressor of invasion and
metastasis in the progression of human bladder cancers. Copyright
(c) 2005 Wiley-Liss, Inc.
PMID: 15880581 [PubMed - indexed for MEDLINE]
9: Neurosci Lett. 2005 Oct 14;387(1):1-4.
Increase of AMPA receptor glutamate receptor 1 subunit and B-cell
receptor-associated protein 31 gene expression in hippocampus of
fatigued mice.
Kamakura M, Tamaki K, Sakaki T, Yoneda Y.
Biotechnology Research Center, Faculty of Engineering, Toyama
Prefectural University, 5180 Kurokawa, Kosugi, Toyama 939-0398, Japan.
kamakura@pu-toyama.ac.jp
Central fatigue is an indispensable biosignal for maintaining life, but
the neuronal and molecular mechanisms involved remain unclear. In this
study, we searched for genes differentially expressed in the
hippocampus of fatigued mice to elucidate the mechanisms underlying
fatigue. Mice were forced to swim in an adjustable-current water pool,
and the maximum swimming time (endurance) until fatigue was measured
thrice. Fatigued and nonfatigued mice with equal swimming capacity and
body weight were compared. We found that the genes of GluR1 and
B-cell receptor-associated protein 31 (Bap31), which acts as a ransport
molecule in the secretory pathway or as a mediator of apoptosis, were
upregulated in the hippocampus of fatigued mice, and increases of GluR1
and Bap31 were confirmed by Northern blotting and real-time PCR. No
change of gene expression of AMPA receptor subunits other than GluR1
was observed. These results suggest that a compositional change of AMPA
receptor (increase of GluR1) and upregulation of the Bap31 gene may be
implicated in fatigue in mice.
PMID: 16051435 [PubMed - indexed for MEDLINE]
10: Science. 2005 Oct 14;310(5746):329-32.
Identification and Functional Characterization of Brainstem Cannabinoid
CB2 Receptors.
Van Sickle MD, Duncan M, Kingsley PJ, Mouihate A, Urbani P, Mackie K,
Stella N, Makriyannis A, Piomelli D, Davison JS, Marnett LJ, Di Marzo
V, Pittman QJ, Patel KD, Sharkey KA.
Institute of Infection, Immunity, and Inflammation and Hotchkiss Brain
Institute, Department of Physiology and Biophysics, University of
Calgary, Calgary, AB, Canada T2N 4N1.
The presence and function of CB(2) receptors in central nervous system
(CNS) neurons are controversial. We report the expression of CB(2)
receptor messenger RNA and protein localization on brainstem neurons.
These functional CB(2) receptors in the brainstem were activated by a
CB(2) receptor agonist, 2-arachidonoylglycerol, and by elevated
endogenous levels of endocannabinoids, which also act at CB(1)
receptors. CB(2) receptors represent an alternative site of action of
endocannabinoids that opens the possibility of nonpsychotropic
therapeutic interventions using enhanced endocannabinoid levels in
localized brain areas.
PMID: 16224028 [PubMed - in process]
11: Eur J Pharmacol. 2005 Oct 12; [Epub ahead of print]
Dose- and duration-dependent effects of betahistine dihydrochloride
treatment on histamine turnover in the cat.
Tighilet B, Trottier S, Lacour M.
UMR 6149 Universite de Provence/CNRS "Neurobiologie Integrative et
Adaptative", Pole 3C << Comportement, Cerveau, Cognition >>, Centre de
St Charles, Case B, 3 Place Victor Hugo, 13331 Marseille Cedex 3,
France.
Drugs interacting with the histaminergic system are currently used for
vertigo treatment and it was shown in animal models that structural
analogues of histamine like betahistine improved the recovery process
after vestibular lesion. This study was aimed at determining the
possible dose and duration effects of betahistine treatment on
histamine turnover in normal adult cats, as judged by the level of
messenger RNA for histidine decarboxylase (enzyme synthesizing
histamine) in the tuberomammillary nuclei. Experiments were
conducted on betahistine-treated cats receiving daily doses of 2, 5,
10, or 50 mg/kg during 1 week, 3 weeks, 2 months, or 3 months. The 1week, 3-week, and 2- and 3-month treatments correspond to the acute,
compensatory, and sustained compensatory stages of vestibular
compensation, respectively. The lowest dose (2 mg/kg) given the longest
time (3 months) was close to the dosage for vestibular defective
patients. Data from the experimental groups were compared to control,
untreated cats and to placebo-treated animals. The results clearly show
that betahistine dihydrochloride administered orally in the normal cat
interferes with histamine turnover by increasing the basal expression
level of histidine decarboxylase mRNA of neurons located in the
tuberomammillary nuclei of the posterior hypothalamus. The effects were
both dose- and time-dependent. In conclusion, compensation of both
static and dynamic deficits is subtended by long-term adaptive
mechanisms that could be facilitated pharmacologically using
betahistine dihydrochloride.
PMID: 16226741 [PubMed - as supplied by publisher]
12: J Biol Chem. 2005 Oct 12; [Epub ahead of print]
Chromosomal instability induced by Pim-1 is passage-dependent and
associated with dysregulation of cyclin B1.
Roh M, Song C, Kim J, Abdulkadir SA.
Pathology Dept., Vanderbilt University, Nashville, TN 37232.
Overexpression of the oncogenic serine/threonine kinase Pim-1 has been
shown to induce chromosomal missegregation and polyploidy in prostate
epithelial cell lines (1). Here we demonstrate that Pim-1-induced
polyploidy develops in a passage-dependent manner in culture consistent
with a stochastic mode of progression. Induction of chromosomal
instability by Pim-1 is not restricted to prostate cells as it was also
observed in telomerase-immortalized normal human mammary epithelial
cells. Elevated levels of cyclin B1 protein, but not its messenger RNA,
were evident in early passage Pim-1 overexpressing cells, suggesting
that increased cyclin B1 levels contribute to the development of
polyploidy. Furthermore, regulation of cyclin B1 protein and cyclin
B1/CDK1 activity after treatment with anti-microtubule agents was
impaired. Small interfering RNA (siRNA) targeting cyclin B1 reversed
the cytokinesis delay but not the mitotic checkpoint defect in Pim-1
overexpressing cells. These results indicate that chronic Pim-1
overexpression dysregulates cyclin B1 protein expression, which
contributes to the development of polyploidy by delaying
cytokinesis.
PMID: 16221667 [PubMed - as supplied by publisher]
13: Vet Immunol Immunopathol. 2005 Oct 11; [Epub ahead of print]
Real-time RT-PCR quantification of mRNA encoding cytokines, CC
chemokines and CCR3 in bronchial biopsies from dogs with eosinophilic
bronchopneumopathy.
Peeters D, Peters IR, Clercx C, Day MJ.
Department of Veterinary Clinical Sciences, Faculty of Veterinary
Medicine, University of Liege, Sart-Tilman B44, 4000 Liege, Belgium.
Idiopathic canine eosinophilic bronchopneumopathy (EBP) is a disease
characterized by eosinophilic infiltration of the pulmonary
interstitium and bronchial mucosa, a cause for which has not yet been
discovered. A recent study, examining the relative proportion of
various lymphocyte cell subsets within bronchoalveolar lavage fluid
from dogs with EBP, has shown a selective increase in CD4(+) T-cells
and a selective decrease in CD8(+) T-cells, suggesting that a
similar Th2 immune response might occur in EBP. The aim of the present
study was to determine the profile of cytokine, chemokine and CC
chemokine receptor 3 (CCR3) messenger RNA (mRNA) expression in
bronchial tissue from dogs with EBP. Real-time RT-PCR assays were used
for the quantification of mRNA encoding for a panel of cytokines, CC
chemokines and CCR3 in perendoscopic bronchial biopsies from eight dogs
with EBP and seven age-matched control dogs. Messenger RNA transcribed
from the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase
was used for normalisation of the threshold cycle in order to determine
the relative copy numbers of the transcripts. No significant difference
in the expression of any cytokine, MCP-1, -2, -4 and CCR3 was found
between control and EBP dogs. The expression of transcript for MCP-3,
eotaxin-2 and -3 was significantly greater in bronchial biopsies from
dogs with EBP than in samples from control dogs while there was
significantly less mRNA encoding RANTES in the mucosa of dogs with EBP.
In conclusion, the cytokine mRNA expression profile in perendoscopic
bronchial biopsies is similar in dogs with EBP and dogs without
respiratory disease. Further studies on the quantification of mRNA
encoding cytokines in isolated T lymphocytes from bronchoalveolar
lavage fluid or bronchial biopsies are needed before any conclusion on
the cytokine profile in canine EBP can be drawn. Eotaxin-2, -3 and MCP3 appear to be implicated in the pathogenesis of the disease.
PMID: 16226318 [PubMed - as supplied by publisher]
14: Neurosci Lett. 2005 Oct 7;386(3):184-8.
Expression profile of addicsin/GTRAP3-18 mRNA in mouse brain.
Inoue K, Akiduki S, Ikemoto MJ.
Age Dimension Research Center, National Institute of Advanced
Industrial Science and Technology, Tsukuba, Ibaraki 305-8566, Japan.
Addicsin is a murine homologue of rat glutamate-transporter-associated
protein 3-18 (GTRAP3-18), a putative modulator of neural glutamatetransporter excitatory amino acid carrier 1 (EAAC1). The other
molecular functions of addicsin, however, remain largely unknown. We
analyzed here the regional and cellular distribution of addicsin
transcript in the mature brain using in situ hybridization analysis,
and examined the sequential addicsin mRNA expression levels in the
developing brain using Northern blot analysis. In the mature brain, we
found addicsin mRNA to be ubiquitously expressed in neuron-like cells,
but not glial cells, in various brain regions including the olfactory
bulbs, hippocampus and cerebral cortex. In the hippocampus, addicsin
mRNA was expressed in the neuron-like cells of the CA1-CA3 pyramidal
cell layer and the interneuron-like cells of the stratum oriens,
stratum radiatum and stratum lacunosum-moleculare. Addicsin transcripts
were also extremely abundant in trigeminal neurons such as the
principal trigeminal nucleus, mesencephalic trigeminal nucleus and
motor trigeminal nucleus. Our evidence suggests that addicsin mRNA is
chiefly expressed in the excitatory and inhibitory neurons, and
that addicsin may participate in the functional expression of the
somatic sensory system by modulation of EAAC1-mediated glutamate
transport. Northern blot analysis revealed that addicsin mRNA levels
increased with maturation, reaching their maximum level at postnatal
day 5 (P5), then significantly declined by about 50% until P14,
suggesting that addicsin may contribute to embryogenesis and
synaptogenesis in the developing brain. Thus, addicsin may
participate in multiple physiological functions in the developing and
mature brain.
PMID: 16005150 [PubMed - indexed for MEDLINE]
*****THROWN OUT – NO ABSTRACT*****
15: Nephrol Dial Transplant. 2005 Oct 4; [Epub ahead of print]
Messenger RNA assessment in clinical nephrology: perspectives and
progress of methodology.
Eikmans M, J Baelde H, de Heer E, A Bruijn J.
Department of Pathology, Leiden University Medical Center, The
Netherlands.
PMID: 16204276 [PubMed - as supplied by publisher]
***** END THROW OUT *****
16: Am J Reprod Immunol. 2005 Oct;54(4):193-202.
Migration inhibitory factor secretion by polarized uterine epithelial
cells is enhanced in response to the TLR3 agonist poly (I:C).
Schaefer TM, Fahey JV, Wright JA, Wira CR.
Department of Physiology, Dartmouth Medical School, 710W Borwell, 1
Medical Center Drive, Lebanon, NH 03756, USA.
todd.m.schaefer@dartmouth.edu
PROBLEM: Uterine epithelial cells produce cytokines that stimulate
leukocytes in response to a microbial insult. The goals of this study
were to determine if uterine epithelial cells produce the proinflammatory cytokine macrophage migration inhibitory factor (MIF), and
to see if toll-like receptor (TLR) agonists stimulate MIF secretion.
METHODS OF STUDY: Human uterine epithelial cells were isolated and
grown in cell culture inserts. Levels of MIF secretion were examined by
ELISA and MIF messenger RNA (mRNA) expression was examined using real
time RT-PCR. RESULTS: Uterine epithelial cells constitutively secrete
MIF and exposure to the TLR3 agonist poly (I:C) resulted in enhanced
apical secretion of MIF. MIF secretion appeared to be from pre-formed
intracellular stores, since exposure of epithelial cells to poly (I:C)
had little effect on the expression of MIF-mRNA. CONCLUSIONS: These
results demonstrate that uterine epithelial cells constitutively
produce MIF and stimulation with poly (I:C) results in enhanced MIF
production. This suggests that MIF secretion by uterine epithelial
cells may play a critical role in innate immune responses against
viral pathogens mediated through TLR3.
PMID: 16135010 [PubMed - in process]
17: Anal Biochem. 2005 Oct 1;345(1):102-9.
mRNA and 18S-RNA coapplication-reverse transcription for quantitative
gene expression analysis.
Zhu LJ, Altmann SW.
Department of Cardiovascular/Metabolic Disease Research, ScheringPlough Research Institute, Kenilworth, NJ 07033, USA.
Fluorescence-based reverse transcription real-time quantitative
polymerase chain reaction (RT-QPCR) is a highly sensitive method for
the detection and quantitation of mRNA. To control and correct for
sample variability, some common housekeeping genes such as
glyceraldehyde-3-phosphate dehydrogenase (GAPDH), beta-actin, and
ubiquitin are often used as endogenous standards. Other internal
calibrators such as 18S-ribosomal RNA (18S-RNA) have also been used,
but further methodological concerns arise given that ribosomal RNA
lacks the 3' poly-A tail typically associated with messenger RNA. To
take advantage of the constant expression levels of 18S-RNA and the
precision of oligo-(dT) primed first-strand synthesis, we have
developed a method that combines oligo-(dT) with an 18S-RNA-specific
primer in the initial reverse transcription (RT) reaction. This
strategy, termed coapplication reverse transcription (Co-RT), allows
for the analysis of multiple target genes with the advantages of 18SRNA normalization from a single RT reaction. In this article, we
describe Co-RT and present tissue distribution and expression level
analysis of several target genes using this method. Co-RT provides
increased sensitivity and higher accuracy than do the standard random
primed RT methods.
PMID: 16139233 [PubMed - in process]
18: Anesthesiology. 2005 Oct;103(4):828-836.
Short Hairpin RNA-mediated Selective Knockdown of NaV1.8 Tetrodotoxinresistant Voltage-gated Sodium Channel in Dorsal Root Ganglion Neurons.
Mikami M, Yang J.
* Postdoctoral Scientist, dagger Professor.
BACKGROUND:: Voltage-gated sodium channels comprise a family of closely
related proteins, each subserving different physiologic and pathologic
functions. NaV1.8 is an isoform of voltage-gated sodium channel
implicated in the pathogenesis of inflammatory and neuropathic pain,
but currently, there is no isoform-specific inhibitor of any voltagegated sodium channels. The authors explored the possibility of short
hairpin RNA-mediated selective knockdown of NaV1.8 expression.
METHODS:: DNA constructs designed to transcribe short hairpin RNA
targeting NaV1.8 were created and incorporated into recombinant
lentiviruses. The virus-induced selective knockdown of NaV1.8 was
examined at the protein, messenger RNA, and functional levels using
Western blot, immunohistochemistry, reverse-transcription polymerase
chain reaction, and patch clamp electrophysiology. RESULTS::
Transduction of HEK293 cells stably expressing NaV1.8 or primary dorsal
root ganglion neurons with lentivirus expressing short hairpin RNA
resulted in the knockdown of NaV1.8 protein and messenger RNA
concentrations. Whole cell patch clamp recordings confirmed decrease in
the NaV1.8-mediated current density without changes in other
biophysical properties.
CONCLUSIONS:: A selective knockdown of NaV1.8 expression in dorsal root
ganglion neurons can be attained by short hairpin RNA delivered with
lentivirus. This method may provide a new gene therapy approach to
controlling neuronal hyperexcitability and pathologic pain.
PMID: 16192776 [PubMed - as supplied by publisher]
19: Arch Neurol. 2005 Oct;62(10):1531-6.
Risk Factors Associated With {beta}-Amyloid(1-42) Immunotherapy in
Preimmunization Gene Expression Patterns of Blood Cells.
O'toole M, Janszen DB, Slonim DK, Reddy PS, Ellis DK, Legault HM, Hill
AA, Whitley MZ, Mounts WM, Zuberek K, Immermann FW, Black RS, Dorner
AJ.
Author Affiliations: Biological Technologies, Wyeth Research,
Cambridge, Mass; Preclinical Biostatistics, Wyeth Research,
Collegeville, Pa.
BACKGROUND: A phase 2a, double-blind, placebo-controlled, multicenter
study was conducted to evaluate safety, tolerability, and pilot
efficacy of immunization with beta-amyloid((1-42)) in patients with
Alzheimer disease. Six immunizations were planned but were halted when
meningoencephalitis was recognized as an adverse event in 6% of
immunized patients. OBJECTIVE: To identify biomarkers associated with
both the risk of meningoencephalitis and antibody responsiveness.
PARTICIPANTS: One hundred fifty-three patients with mild to
moderate Alzheimer disease. Main Outcome Measure Association between
response to immunization and preimmunization expression levels of 8239
messenger RNA transcripts expressed in peripheral blood mononuclear
cells that had been collected at the screening visit. RESULTS:
Expression patterns of genes related to apoptosis and proinflammatory
pathways (tumor necrosis factor pathway in particular) were identified
as biomarkers of risk for the development of meningoencephalitis.
Expression patterns of genes related to protein synthesis,
protein trafficking, DNA recombination, DNA repair, and cell cycle were
strongly associated with IgG response to immunization. CONCLUSIONS:
Candidate biomarkers associated with risk of immunotherapy-related
meningoencephalitis were detected in blood collected prior to
treatment. In addition, a different set of biomarkers were identified
that were associated with the desired outcome of IgG response.
PMID: 16216935 [PubMed - in process]
20: Arthritis Rheum. 2005 Oct;52(10):3257-68.
Synovial fibroblasts promote osteoclast formation by RANKL in a novel
model of spontaneous erosive arthritis.
Wu Y, Liu J, Feng X, Yang P, Xu X, Hsu HC, Mountz JD.
University of Alabama at Birmingham, Birmingham, Alabama.
OBJECTIVE: Erosion of cartilage and bone is a hallmark of rheumatoid
arthritis (RA). This study was undertaken to explore the roles of
hyperproliferating synovial fibroblasts and macrophages in abnormal
osteoclast formation, using the recently described BXD2 mouse model of
RA. METHODS: Cell distribution in the joints was analyzed by
immunohistochemistry, using tartrate-resistant acid phosphatase (TRAP)
staining to identify osteoclasts. To identify the defective
cells in BXD2 mice, mouse synovial fibroblasts (MSFs) were cultured
with bone marrow-derived macrophages. Osteoclast formation was assayed
by TRAP staining and bone resorption pit assay, and the cytokine
profiles of the MSFs and macrophages were determined by quantitative
real-time polymerase chain reaction and enzyme-linked immunosorbent
assay. RESULTS: In BXD2 mice, TRAP-positive osteoclasts were found at
sites of active bone erosion, in close proximity to hyperproliferating
synovial fibroblasts. On coculture, MSFs from BXD2 mice, but
not C57BL/6 mice, produced high levels of RANKL messenger RNA, induced
macrophages to form osteoclasts, and actively eroded bone slices,
through a mechanism(s) that could be blocked by pretreatment with
osteoprotegerin. Although macrophages from BXD2 mice expressed higher
basal levels of tumor necrosis factor alpha (TNFalpha), interleukin1beta (IL-1beta), and IL-6 than those from C57BL/6 mice, abnormal
osteoclast formation was not due to enhanced sensitivity of the BXD2
mouse macrophages to RANKL. TNFalpha, produced by both BXD2 MSFs and
BXD2 mouse macrophages, had a strong stimulatory effect on RANKL
expression. CONCLUSION: BXD2 MSFs produce RANKL and induce the
development of osteoclasts from macrophages. The enhanced production of
RANKL is possibly due to autocrine stimulation, together with paracrine
stimulation by factors produced by macrophages.
PMID: 16200600 [PubMed - in process]
21: Biol Reprod. 2005 Oct;73(4):840-7. Epub 2005 Jun 29.
Meiotic Messenger RNA and Noncoding RNA Targets of the RNA-Binding
Protein Translin (TSN) in Mouse Testis.
Cho YS, Iguchi N, Yang J, Handel MA, Hecht NB.
Center for Research on Reproduction and Women's Health, University of
Pennsylvania School of Medicine, Philadelphia, Pennsylvania 19104.
In postmeiotic male germ cells, TSN, formerly known as testis brain-RNA
binding protein, is found in the cytoplasm and functions as a
posttranscriptional regulator of a group of genes transcribed by the
transcription factor CREM-tau. In contrast, in pachytene spermatocytes,
TSN is found predominantly in nuclei. Tsn-null males show a reduced
sperm count and high levels of apoptosis in meiotic cells, suggesting a
critical function for TSN during meiosis. To identify meiotic target
RNAs that associate in vivo with TSN, we reversibly cross-linked TSN to
RNA in testis extracts from 17-day-old and adult mice and
immunoprecipitated the complexes with an affinity-purified TSN
antibody. Extracts from Tsn-null mice were used as controls. Cloning
and sequencing the immunoprecipitated RNAs, we identified four new TSN
target mRNAs, encoding diazepam-binding inhibitor-like 5, arylsulfatase
A, a tetratricopeptide repeat structure-containing protein, and ring
finger protein 139. In contrast to the population of postmeiotic
translationally delayed mRNAs that bind TSN, these four mRNAs are
initially expressed in pachytene spermatocytes. In addition,
anti-TSN also precipitated a nonprotein-coding RNA (ncRNA), which is
abundant in nuclei of pachytene spermatocytes and has a putative
polyadenylation signal, but no open reading frame. A second similar
ncRNA is adjacent to a GGA repeat, a motif frequently associated with
recombination hot spots. RNA gel-shift assays confirm that the four new
target mRNAs and the ncRNA specifically bind to TSN in testis extracts.
These studies have, for the first time, identified both mRNAs and a
ncRNA as TSN targets expressed during meiosis.
PMID: 15987823 [PubMed - in process]
22: Cell Stress Chaperones. 2005 Autumn;10(3):204-10.
Expression of mRNA for the t-complex polypeptide-1, a subunit of
chaperonin CCT, is upregulated in association with increased cold
hardiness in Delia antiqua.
Kayukawa T, Chen B, Miyazaki S, Itoyama K, Shinoda T, Ishikawa Y.
Laboratory of Applied Entomology, Graduate School of Agricultural and
Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Summer-diapause and winter-diapause pupae of the onion maggot, Delia
antique (Diptera: Anthomyiidae), were significantly more cold hardy
than nondiapause, prediapause, and postdiapause pupae. Moreover, cold
acclimation of nondiapause pupae conferred strong cold hardiness
comparable with that of diapause pupae. Differential display analysis
revealed that the expression of a gene encoding TCP-1 (the t-complex
polypeptide-1), a subunit of chaperonin CCT, in D antique (DaTCP-1) is
upregulated in the pupae that express enhanced cold hardiness.
Quantitative real-time polymerase chain reaction analyses showed that
the levels of DaTCP-1 messenger RNA in pupal tissues, brain, and midgut
in particular, are highly correlated with the cold hardiness of the
pupae. These findings suggest that the upregulation of DaTCP-1
expression is related to enhanced cold hardiness in D antiqua. The
upregulation of CCT in response to low temperature in an organism other
than the yeast is newly reported.
PMID: 16184765 [PubMed - in process]
23: Crit Care Med. 2005 Oct;33(10):2323-2331.
Carbon monoxide, but not endothelin-1, plays a major role for the
hepatic microcirculation in a murine model of early systemic
inflammation.
Wunder C, Brock RW, Frantz S, Gottsch W, Morawietz H, Roewer N,
Eichelbronner O.
From Klinik und Poliklinik fur Anasthesiologie (CW, NR, OE) and
Medizinische Klinik (SF), Julius-Maximilians-Universitat Wurzburg,
Wurzburg, Germany; Department of Pharmacology and Toxicology,
University of Arkansas of Medical Sciences, Little Rock, AR (RWB); and
Medizinische Klinik, Universitatsklinikum Carl Gustav Carus, Technical
University Dresden, Dresden, Germany (WG, HM).
OBJECTIVE:: Endothelin-1 and carbon monoxide play a major role in the
regulation of liver microcirculation in numerous disease states. During
sepsis and endotoxemia, elevated formation of endothelin-1 results in
reduced sinusoidal blood flow. However, the role of carbon monoxide and
endothelin-1 and its receptors endothelin receptor A and endothelin
receptor B in the deranged liver microcirculation during early systemic
inflammation remains unclear. DESIGN:: Prospective, randomized,
controlled experiment. SETTING:: University animal laboratory.
SUBJECTS:: Male C57/BL6 mice, weighing 23-27 g. INTERVENTIONS:: To
induce a systemic inflammation, mice were treated with 1 hr of
bilateral hind limb ischemia followed by 3 hrs or 6 hrs of reperfusion.
Animals were randomly exposed to the nonselective endothelin receptor
antagonist Ro-61-6612 (Tezosentan) and/or a continuous endothelin-1
infusion. Different animals were randomized to methylene chloride
gavage or carbon monoxide inhalation during the reperfusion period.
MEASUREMENTS AND MAIN RESULTS:: After ischemia/reperfusion, endothelin1 plasma concentrations, endothelin-1 messenger RNA expression, and
endothelin receptor A and B messenger RNA expression revealed no
significant changes when compared with sham animals. After 6 hrs of
ischemia/reperfusion, hepatic microcirculatory variables (sinusoidal
density, sinusoidal diameter, and red blood cell velocity)
deteriorated. Tezosentan after 6 hrs of ischemia/reperfusion did not
improve the liver microcirculation, whereas the continuous infusion of
endothelin-1 after 6 hrs of ischemia/reperfusion further impaired
sinusoidal blood flow. Tezosentan treatment did not produce any
alterations in hepatocellular injury or hepatic redox status when
compared with the untreated animals receiving 6 hrs of
schemia/reperfusion. Animals receiving 6 hrs of ischemia/reperfusion
and exposed to methylene chloride gavage or inhaled carbon monoxide
during limb reperfusion showed significantly improved microcirculatory
variables, hepatic redox status, and attenuated hepatocellular
injury. CONCLUSIONS:: These data suggest that endothelin-1 and the
endothelin receptors A and B are not responsible for the observed
hepatic microcirculatory and cellular dysfunction during early systemic
inflammation, but exposure to exogenous carbon monoxide protected the
hepatic microcirculation and improved the impaired hepatic cellular
integrity and the hepatocellular redox status.
PMID: 16215388 [PubMed - as supplied by publisher]
24: Crit Care Med. 2005 Oct;33(10):2309-2316.
Rosiglitazone, a peroxisome proliferator-activated receptor-gamma
agonist, reduces acute lung injury in endotoxemic rats.
Liu D, Zeng BX, Zhang SH, Wang YL, Zeng L, Geng ZL, Zhang SF.
From the Department of Anesthesiology (DL, BXZ, SHZ, YLW, LZ), Union
Hospital, Tongji Medical College, Huazhong University of Science and
Technology, Wuhan, China; and the Department of Anesthesiology (ZLG)
and Department of Thoracic and Cardiac Surgery (SFZ), Lanzhou General
Hospital of P.L.A., Lanzhou, China.
OBJECTIVE:: Rosiglitazone, a potent agonist of peroxisome proliferatoractivated receptor (PPAR)-gamma, exerts anti-inflammatory effects in
vitro and in vivo. This study was designated to determine the effects
of rosiglitazone on endotoxin-induced acute lung injury in rats.
DESIGN:: Prospective, experimental study. SETTING:: University research
laboratory. SUBJECTS:: Thirty-six male Wistar rats. INTERVENTIONS:: All
the animals were randomly assigned to one of six groups (n = 6 per
group) and were given either lipopolysaccharide (6 mg/kg intravenously)
or saline, pretreated with rosiglitazone (0.3 mg/kg intravenously) or
vehicle (10% dimethyl sulphoxide) 30 mins before lipopolysaccharide.
The selective PPAR-gamma antagonist GW9662 (0.3 mg/kg intravenously) or
its vehicle (10% dimethyl sulphoxide) was given 20 mins before
rosiglitazone. MEASUREMENTS AND MAIN RESULTS:: Endotoxemia for 4 hrs
induced evident lung histologic injury and edema, both of which were
significantly attenuated by rosiglitazone pretreatment. The protective
effects of rosiglitazone were correlated with the reduction by 71% of
the increase of myeloperoxidase activity and the reduction by 84% of
the increase of malondialdehyde in the lung tissue. The pulmonary
hyperproduction of nitric oxide was reduced by 82% of the increase
related to lipopolysaccharide challenge. Pretreatment with
rosiglitazone also markedly suppressed lipopolysaccharide-induced
expression of inducible nitric oxide synthase messenger RNA and protein
in the lung, as demonstrated by reverse transcription-polymerase chain
reaction or Western blot analysis. Immunohistochemical analysis
revealed that rosiglitazone inhibited the formation of nitrotyrosine, a
marker for peroxynitrite reactivity, in the lung tissue. In
addition, the specific PPAR-gamma antagonist GW9662 antagonized the
effects of rosiglitazone. CONCLUSIONS:: This study provides evidence,
for the first time, that the PPAR-gamma agonist rosiglitazone
significantly reduces endotoxin-induced acute lung injury in rats.
PMID: 16215386 [PubMed - as supplied by publisher]
25: EMBO Rep. 2005 Oct;6(10):968-72.
Calicivirus translation initiation requires an interaction between VPg
and eIF4E.
Goodfellow I, Chaudhry Y, Gioldasi I, Gerondopoulos A, Natoni A, Labrie
L, Laliberte JF, Roberts L.
School of Animal and Microbial Sciences, University of Reading,
Whiteknights, Reading RG6 6AJ, UK.
Unlike other positive-stranded RNA viruses that use either a 5'-cap
structure or an internal ribosome entry site to direct translation of
their messenger RNA, calicivirus translation is dependent on the
presence of a protein covalently linked to the 5' end of the viral
genome (VPg). We have shown a direct interaction of the calicivirus VPg
with the cap-binding protein eIF4E. This interaction is required for
calicivirus mRNA translation, as sequestration of eIF4E by 4E-BP1
inhibits translation. Functional analysis has shown that VPg
does not interfere with the interaction between eIF4E and the cap
structure or 4E-BP1, suggesting that VPg binds to eIF4E at a different
site from both cap and 4E-BP1. This work lends support to the idea that
calicivirus VPg acts as a novel 'cap substitute' during initiation of
translation on virus mRNA.
PMID: 16142217 [PubMed - in process]
26: Fertil Steril. 2005 Oct;84 Suppl 2:1095-103.
Expression of human oviductin in an immortalized human oviductal cell
line.
Ling L, Lee YL, Lee KF, Tsao SW, Yeung WS, Kan FW.
Department of Anatomy and Cell Biology, Faculty of Health Sciences,
Queen's University, Kingston, Ontario, Canada.
OBJECTIVE: To determine whether OE-E6/E7, an immortalized human
oviductal epithelial cell line, expresses oviductin messenger RNA
(mRNA) and its translated protein. DESIGN: Transmission electron
microscopy was employed to characterize the morphology of OE-E6/E7
cells followed by reverse-transcription polymerase chain reaction (PCR)
analysis of oviductin mRNA and sequencing of the nested-PCR product.
Confocal microscopy was used, using a polyclonal antibody against human
oviductin and Con A as a marker for mannose residues, to reveal the
colocalization of human oviduct-specific glycoprotein with the
endoplasmic reticulum and Golgi compartments. SETTING: University-based
anatomy and cell biology department. PATIENT(S): Women undergoing
laparoscopy for tubal ligation or hysterectomy due to uterine fibroma.
INTERVENTION(S): An immortalized OE-E6/E7 cell line was previously
established using human oviductal epithelial cells. Electron
microscopy, RT-PCR, sequencing, immunohistochemistry and confocal
microscopy were performed. MAIN OUTCOME MEASURE(S): The presence of
human oviductin mRNA and protein in OE-E6/E7 cells. RESULT(S): OE-E6/E7
cells retain morphological features characteristic of secretory cells
and express human oviductin mRNA and its translated protein.
CONCLUSION(S): OE-E6/E7 cells were characterized for the first time by
electron microscopy and shown to exhibit histological features typical
of secretory cells. Reverse-transcription PCR with sequencing and
confocal microscopy showed, respectively, that human oviductin mRNA and
protein are expressed in OE-E6/E7 cells. Our results suggest that OEE6/E7 could be a useful tool for future studies of the function of
human oviductin.
PMID: 16209999 [PubMed - in process]
27: Gynecol Oncol. 2005 Oct;99(1):183-8.
A-kinase anchoring protein 3 messenger RNA expression correlates with
poor prognosis in epithelial ovarian cancer.
Sharma S, Qian F, Keitz B, Driscoll D, Scanlan MJ, Skipper J, Rodabaugh
K, Lele S, Old LJ, Odunsi K.
Department of Gynecologic Oncology, Roswell Park Cancer Institute, Elm
and Carlton Streets, Buffalo, NY 14263, USA.
OBJECTIVES: Cancer-testis (CT) antigens are expressed in tumors but not
normal tissues except the testis and could be targets for vaccine
therapy in epithelial ovarian cancer (EOC). A-kinase anchoring protein
3 (AKAP-3) is a novel member of the CT antigen family. The aim of this
study was to examine the expression of AKAP-3 in EOC and correlate with
clinico-pathologic characteristics. METHODS: One step RT-PCR was
performed with RNA from normal and ovarian cancer cell lines
and 74 epithelial ovarian tumor tissues. AKAP-3-specific PCR product
was amplified. The distribution of AKAP-3 expression and clinicopathologic variables was analyzed. Survival distributions were
estimated, and multivariate analyses were performed. RESULTS: AKAP-3
mRNA expression was demonstrated in 43/74 (58%) of the ovarian cancer
specimens. AKAP-3 was expressed in normal testis, but not in other
normal tissues. AKAP-3 expression significantly correlated with
increased likelihood of residual tumor (P = 0.005), but no increase in
the likelihood of recurrence or persistent disease (P = 0.06).
Patients with AKAP-3 mRNA expression were found to have a significantly
poorer overall survival (median 50 months) compared with patients
without AKAP-3 expression (median not reached) (P = 0.007).
Multivariate analysis of AKAP-3 expression, residual disease, and
response to frontline chemotherapy found response to be the strongest
predictor of overall survival (P = 0.012).
CONCLUSIONS: Our data demonstrate that AKAP-3 is expressed at high
frequency in patients with EOC. Since AKAP-3 demonstrates tumorrestricted expression and appears to be associated with worse overall
survival, it could represent an attractive target for antigen-specific
immunotherapy in EOC.
PMID: 16005946 [PubMed - in process]
28: Hum Pathol. 2005 Oct;36(10):1037-48. Epub 2005 Sep 6.
Increased expression of delta-catenin/neural plakophilin-related
armadillo protein is associated with the down-regulation and
redistribution of E-cadherin and p120(ctn) in human prostate cancer.
Lu Q, Dobbs LJ, Gregory CW, Lanford GW, Revelo MP, Shappell S, Chen YH.
Department of Anatomy and Cell Biology, Brody School of Medicine, East
Carolina University, Greenville, NC 27858, USA.
delta-Catenin, or neural plakophilin-related armadillo protein, is a
unique armadillo domain-containing protein in that it is neuralspecific and primarily expressed in the brain. However, our recent
analysis of the human genome revealed a consistent association of
delta-catenin messenger RNA sequences with malignant cells, although
the significance of these findings was unclear. In this study, we
report that a number of delta-catenin epitopes were expressed in
human prostate cancer cells. Western blot and tissue microarray
revealed a close association between increased delta-catenin expression
and human primary prostatic adenocarcinomas. The analyses of 90 human
prostate cancer and 90 benign prostate tissue samples demonstrated that
an estimated 85% of prostatic adenocarcinomas showed enhanced deltacatenin immunoreactivity. delta-Catenin expression increased with
prognostically significant increased Gleason scores. By analyzing the
same tumor cell clusters using consecutive sections, we showed
that an increased delta-catenin immunoreactivity was accompanied by the
down-regulation and redistribution of E-cadherin and p120(ctn), major
cell junction proteins whose inactivation is frequently associated with
cancer progression. Furthermore, overexpression of delta-catenin in
tumorigenic CWR-R1 cells that are derived from human prostate cancer
xenograft resulted in reduced immunoreactivity for E-cadherin and
p120(ctn) at the cell-cell junction. This is the first study comparing
overexpression of delta-catenin with the E-cadherin/catenin system in
cancer and shows that delta-catenin may be intimately involved in
regulating E-cadherin/p120(ctn) cell-cell adhesion in prostate cancer
progression.
PMID: 16226102 [PubMed - in process]
29: Int J Biochem Cell Biol. 2005 Oct;37(10):2156-68. Epub 2005 Mar 13.
Novel aspects on the regulation of muscle wasting in sepsis.
Hasselgren PO, Menconi MJ, Fareed MU, Yang H, Wei W, Evenson A.
Department of Surgery, Beth Israel Deaconess Medical Center, Harvard
Medical School, 330 Brookline Avenue, Boston, MA 02215, USA.
phasselg@bidmc.harvard.edu
Muscle wasting in sepsis is associated with increased expression of
messenger RNA for several genes in the ubiquitin-proteasome proteolytic
pathway, indicating that increased gene transcription is involved in
the development of muscle atrophy. Here we review the influence of
sepsis on the expression and activity of the transcription factors
activator protein-1, nuclear factor-kappaB (NF-kappaB), and
CCAAT/enhancer binding protein, as well as the nuclear cofactor
p300. These transcription factors may be important for sepsis-induced
muscle wasting because several of the genes in the ubiquitin-proteasome
proteolytic pathway have multiple binding sites for activating protein1, nuclear factor-kappaB, and CCAAT/enhancer binding protein in their
promoter regions. In addition, the potential role of increased muscle
calcium levels for sepsis-induced muscle atrophy is reviewed. Calcium
may regulate several mechanisms and factors involved in muscle wasting,
including the expression and activity of the calpain-calpastatin
system, proteasome activity, CCAAT/enhancer binding protein
transcription factors, apoptosis and glucocorticoid-mediated
muscle protein breakdown. Because muscle wasting is commonly seen in
patients with sepsis and has severe clinical consequences, a better
understanding of mechanisms regulating sepsis-induced muscle wasting
may help improve the care of patients with sepsis and other musclewasting conditions as well.
PMID: 16125115 [PubMed - in process]
30: Int J Hematol. 2005 Oct;82(3):224-9.
Arsenic Trioxide Therapy in Relapsed or Refractory Japanese Patients
with Acute Promyelocytic Leukemia: Updated Outcomes of the Phase II
Study and Postremission Therapies.
Shigeno K, Naito K, Sahara N, Kobayashi M, Nakamura S, Fujisawa S,
Shinjo K, Takeshita A, Ohno R, Ohnishi K.
Department of Medicine III, Hamamatsu University School of Medicine,
Hamamatsu.
Recently, arsenic trioxide (ATO) has been proved to induce complete
remission (CR) at a high rate in patients with acute promyelocytic
leukemia (APL).We prospectively investigated the safety and efficacy of
ATO therapy in patients with relapsed and refractory APL and examined
the duration of CR and the postremission therapies. Initially, 0.15
mg/kg ATO was administered until bone marrow remission to a maximum of
60 days. After the patient achieved CR, 1 additional ATO course at the
same dosage was administered for 25 days. Of 34 patients, 31 (91%)
achieved CR. PML-RAR3 messenger RNA was not detected in the bone marrow
of 18 (72%) of the 25 patients evaluated by reverse transcriptasepolymerase chain reaction analysis. At a median follow-up of 30
months, the estimated 2-year overall survival rate was 56%, and the
estimated 2-year event-free survival rate was 17%. During the ATO
therapy, QTc prolongation was observed in most cases. Fifteen patients
developed ventricular tachycardia, and 1 of them showed torsades de
pointes. Other adverse events were nausea, water retention, APL
differentiation syndrome, skin eruption, liver dysfunction, and
peripheral neuropathy, all of which were quite tolerable. ATO
therapy was remarkably effective for relapsed APL; however,
postremission therapies were necessary to maintain a durable remission.
PMID: 16207595 [PubMed - in process]
31: Int J Hematol. 2005 Oct;82(3):215-23.
The Expression of Telomeric Proteins and Their Probable Regulation of
Telomerase during the Differentiation of All-trans-Retinoic AcidResponsive and –Resistant Acute Promyelocytic Leukemia Cells.
Sun J, Huang H, Zhu Y, Lan J, Li J, Lai X, Yu J.
Department of Hematology, College of Medicine, Zhejiang University,
Hangzhou, China.
Telomerase activity has been linked to retinoid induction of tumor cell
differentiation, and the patterns of telomerase expression are
different in the 2 pathways of acute promyelocytic leukemia (APL) cell
differentiation: the retinoic acid receptor 3 (RAR3)-dependent and the
retinoic X receptor 3 (RXR3)-dependent pathways. Still, whether
telomeric proteins respond to retinoid treatment is not clear. If they
do, how they would respond and how they would interfere in telomerase
regulation during differentiation are also unclear. Using all-transretinoic acid (ATRA)-sensitive and -resistant APL cell lines NB4, NB4R1, and NB4-R2, we analyzed a panel of telomeric proteins, including
TRF1, PINX1, TANK1, and TANK2, at the messenger RNA (mRNA) and protein
expression levels during the differentiation of these cell lines in the
2 pathways. Our analyses showed that both mRNA and protein expression
of TRF1 remained stable during NB4 and NB4-R1 cell differentiation but
slightly increased in NB4-R2 cells, suggesting that TRF1 may have
different functions in the RAR3- and RXR3-dependent pathways. The
stable expression of TRF1 may be because telomere length remains
unchanged. Pinx1 mRNA expression was tightly correlated with telomerase
reverse transcriptase (hTERT) mRNA expression during differentiation.
Variation in Pinx1 expression may be a reaction induced by
hTERT expression variation. TANK1 mRNA expression and TANK1 protein
levels were both down-regulated in all 3 APL cell lines at a later
period of differentiation, suggesting that TANK1 may positively
regulate telomerase activity and that both RAR3- and RXR3-dependent
pathways may exert this regulation.TANK2 expression levels remained
stable in all 3 APL cell lines during differentiation, showing that
TANK2 may have little effect on telomerase. Thus, our studies provide
an outline of the dynamics of telomeric protein expression and the
probable regulatory effects of these proteins on telomerase during the
differentiation of ATRA-responsive and -resistant APL cells.
PMID: 16207594 [PubMed - in process]
Keyword search: “Messenger RNA”
1: Mol Genet Genomics. 2005 Oct 18;:1-11 [Epub ahead of print]
DEG1, encoding the tRNA:pseudouridine synthase Pus3p, impacts HOT1stimulated recombination in Saccharomyces cerevisiae.
Hepfer CE, Arnold-Croop S, Fogell H, Steudel KG, Moon M, Roff A,
Zaikoski S, Rickman A, Komsisky K, Harbaugh DL, Lang GI, Keil RL.
Department of Biology, Millersville University, 50 East Frederick
Street, PO Box 1002, Millersville, PA, 17551, USA,
carol.hepfer@millersville.edu.
In Saccharomyces cerevisiae, HOT1-stimulated recombination has been
implicated in maintaining homology between repeated ribosomal RNA
genes. The ability of HOT1 to stimulate genetic exchange requires RNA
polymerase I transcription across the recombining sequences. The transacting nuclear mutation hrm3-1 specifically reduces HOT1-dependent
recombination and prevents cell growth at 37 degrees . The HRM3 gene is
identical to DEG1. Excisive, but not gene replacement, recombination is
reduced in HOT1-adjacent sequences in deg1Delta mutants. Excisive
recombination within the genomic rDNA repeats is also decreased. The
hypo-recombination and temperature-sensitive phenotypes of deg1Delta
mutants are recessive. Deletion of DEG1 did not affect the rate of
transcription from HOT1 or rDNA suggesting that while transcription is
necessary it is not sufficient for HOT1 activity. Pseudouridine
synthase 3 (Pus3p), the DEG1 gene product, modifies the anticodon arm
of transfer RNA at positions 38 and 39 by catalyzing the conversion of
uridine to pseudouridine. Cells deficient in pseudouridine synthases
encoded by PUS1, PUS2 or PUS4 displayed no recombination defects,
indicating that Pus3p plays a specific role in HOT1 activity. Pus3p is
unique in its ability to modulate frameshifting and readthrough events
during translation, and this aspect of its activity may be responsible
for HOT1 recombination phenotypes observed in deg1 mutants.
PMID: 16231152 [PubMed - as supplied by publisher]
2: Nat Struct Mol Biol. 2005 Oct 16; [Epub ahead of print]
Structural perspective on the activation of RNase P RNA by protein.
Buck AH, Kazantsev AV, Dalby AB, Pace NR.
Department of Chemistry and Biochemistry, University of Colorado,
Boulder, Colorado 80309, USA.
Ribonucleoprotein particles are central to numerous cellular pathways,
but their study in vitro is often complicated by heterogeneity and
aggregation. We describe a new technique to characterize these
complexes trapped as homogeneous species in a nondenaturing gel. Using
this technique, in conjunction with phosphorothioate footprinting
analysis, we identify the protein-binding site and RNA folding states
of ribonuclease P (RNase P), an RNA-based enzyme that, in
vivo, requires a protein cofactor to catalyze the 5' maturation of
precursor transfer RNA (pre-tRNA). Our results show that the protein
binds to a patch of conserved RNA structure adjacent to the active site
and influences the conformation of the RNA near the tRNA-binding site.
The data are consistent with a role of the protein in substrate
recognition and support a new model of the holoenzyme that is based on
a recently solved crystal structure of RNase P RNA.
PMID: 16228004 [PubMed - as supplied by publisher]
3: Chembiochem. 2005 Oct 5; [Epub ahead of print]
Mechanism and Substrate Specificity of tRNA-Guanine Transglycosylases
(TGTs): tRNA-Modifying Enzymes from the Three Different Kingdoms of
Life Share a Common Catalytic Mechanism.
Stengl B, Reuter K, Klebe G.
Institut fur Pharmazeutische Chemie Philipps-Universitat Marburg
Marbacher Weg 6, 35032 Marburg, Germany, Fax: (+49) 6421-282-8994.
Transfer RNA-guanine transglycosylases (TGTs) are evolutionarily
ancient enzymes, present in all kingdoms of life, catalyzing guanine
exchange within their cognate tRNAs by modified 7-deazaguanine bases.
Although distinct bases are incorporated into tRNA at different
positions in a kingdom-specific manner, the catalytic subunits of TGTs
are structurally well conserved. This review provides insight into the
sequential steps along the reaction pathway, substrate specificity, and
conformational adaptions of the binding pockets by comparison of TGT
crystal structures in complex with RNA substrates of a eubacterial and
an archaebacterial species. Substrate-binding modes indicate an
evolutionarily conserved base-exchange mechanism with a conserved
aspartate serving as a nucleophile through covalent binding to C1' of
the guanosine ribose moiety in an intermediate state. A second
conserved aspartate seems to control the spatial rearrangement of the
ribose ring along the reaction pathway and supposedly operates as a
general acid/base. Water molecules inside the binding pocket
accommodating interaction sites subsequently occupied by polar atoms of
substrates help to elucidate substrate-recognition and substratespecificity features. This emphasizes the role of water molecules as
general probes to map binding-site properties for structure-based drug
design. Additionally, substrate-bound crystal structures allow the
extraction of valuable information about the classification of the TGT
superfamily into a subdivision of presumably homologous superfamilies
adopting the triose-phosphate isomerase type barrel fold with a
standard phosphate-binding motif.
PMID: 16206323 [PubMed - as supplied by publisher]
4: Mar Biotechnol (NY). 2005 Sep 28; [Epub ahead of print]
Mitochondrial DNA Sequence and Gene Organization in Australian Backup
Abalone Haliotis Rubra (Leach).
Maynard BT, Kerr LJ, McKiernan JM, Jansen ES, Hanna PJ.
School of Biological & Chemical Sciences, Deakin University, Geelong,
VIC 3217, Australia, pjh@deakin.edu.au.
The complete mitochondrial DNA of the blacklip abalone Haliotis rubra
(Gastropoda: Mollusca) was cloned and 16,907 base pairs were sequenced.
The sequence represents an estimated 99.85% of the mitochondrial
genome, and contains 2 ribosomal RNA, 22 transfer RNA, and 13 proteincoding genes found in other metazoan mtDNA. An AT tandem repeat and a
possible C-rich domain within the putative control region could not be
fully sequenced. The H. rubra mtDNA gene order is novel for mollusks,
separated from the black chiton Katharina tunicata by the individual
translocations of 3 tRNAs. Compared with other mtDNA regions, sequences
from the ATP8, NAD2, NAD4L, NAD6, and 12S rRNA genes, as well as the
control region, are the most variable among representatives from
Mollusca, Arthropoda, and Rhynchonelliformea, with similar mtDNA
arrangements to H. rubra. These sequences are being evaluated as
genetic markers within commercially important Haliotis species, and
some applications and considerations for their use are discussed.
PMID: 16206015 [PubMed - as supplied by publisher]
5: Nucleic Acids Res. 2005 Oct 4;33(17):5544-52. Print 2005.
Competition between trans-translation and termination or elongation of
translation.
Asano K, Kurita D, Takada K, Konno T, Muto A, Himeno H.
Department of Biochemistry and Biotechnology, Faculty of Agriculture
and Life Science, Hirosaki University, Hirosaki 036-8561, Japan.
The effects of tRNA, RF1 and RRF on trans-translation by tmRNA were
examined using a stalled complex of ribosome prepared using a synthetic
mRNA and pure Escherichia coli translation factors. No
endoribonucleolytic cleavage of mRNA around the A site was found in the
stalled ribosome and was required for the tmRNA action. When the A site
was occupied by a stop codon, alanyl-tmRNA competed with RF1 with the
efficiency of peptidyl-transfer to alanyl-tmRNA for trans-translation
inversely correlated to the efficiency of translation termination. The
competition was not affected by RF3. A sense codon also serves
as a target for alanyl-tmRNA with competition of aminoacyl-tRNA. The
extent of inhibition was decreased with the length of the 3'-extension
of mRNA. RRF, only at a high concentration, slightly affected peptidyltransfer for trans-translation, although it did not affect the
canonical elongation. These results indicate that alanyl-tmRNA does not
absolutely require the truncation of mRNA around the A site but prefers
an mRNA of a short 3'-extension from the A site and that it can operate
on either a sense or termination codon at the A site, at which alanyltmRNA competes with aminoacyl-tRNA, RF and RRF.
PMID: 16204455 [PubMed - indexed for MEDLINE]
6: Nucleic Acids Res. 2005 Sep 30;33(17):5647-58. Print 2005.
A new mechanism for mtDNA pathogenesis: impairment of posttranscriptional maturation leads to severe depletion of mitochondrial
tRNASer(UCN) caused by T7512C and G7497A point mutations.
Mollers M, Maniura-Weber K, Kiseljakovic E, Bust M, Hayrapetyan A,
Jaksch M, Helm M, Wiesner RJ, von Kleist-Retzow JC.
Institute of Vegetative Physiology, University of Koln, Robert-KochStrasse 39, 50931 Koln, Germany.
We have studied the consequences of two homoplasmic, pathogenic point
mutations (T7512C and G7497A) in the tRNA(Ser(UCN)) gene of
mitochondrial (mt) DNA using osteosarcoma cybrids. We identified a
severe reduction of tRNA(Ser(UCN)) to levels below 10% of controls for
both mutations, resulting in a 40% reduction in mitochondrial protein
synthesis rate and in a respiratory chain deficiency resembling that in
the patients muscle. Aminoacylation was apparently unaffected. On nondenaturating northern blots we detected an altered electrophoretic
mobility for G7497A containing tRNA molecules suggesting a structural
impact of this mutation, which was confirmed by structural probing.
By comparing in vitro transcribed molecules with native RNA in such
gels, we also identified tRNA(Ser(UCN)) being present in two isoforms
in vivo, probably corresponding to the nascent, unmodified transcripts
co-migrating with the in vitro transcripts and a second, faster moving
isoform corresponding to the mature tRNA. In cybrids containing either
mutations the unmodified isoforms were severely reduced. We hypothesize
that both mutations lead to an impairment of post-transcriptional
modification processes, ultimately leading to a preponderance of
degradation by nucleases over maturation by modifying enzymes,
resulting in severely reduced tRNA(Ser(UCN)) steady state levels. We
infer that an increased degradation rate, caused by disturbance of tRNA
maturation and, in the case of the G7497A mutant, alteration of tRNA
structure, is a new pathogenic mechanism of mt tRNA point mutations.
PMID: 16199753 [PubMed - indexed for MEDLINE]
*****THROWN OUT – NO ABSTRACT*****
7: FASEB J. 2005 Oct;19(12):1583-4.
50 years ago protein synthesis met molecular biology: the discoveries
of amino acid activation and transfer RNA.
Pederson T.
Department of Biochemistry and Molecular Pharmacology, University of
Massachusetts Medical School, Worcester, Massachusetts, USA.
thoru.pederson@umassmed.edu
PMID: 16195365 [PubMed - in process]
***** END THROW OUT *****
8: Appl Microbiol Biotechnol. 2005 Sep 29;:1-10 [Epub ahead of print]
Secreted beta-galactosidase from a Flavobacterium sp. isolated from a
low-temperature environment.
Sorensen HP, Porsgaard TK, Kahn RA, Stougaard P, Mortensen KK, Johnsen
MG.
Laboratory of BioDesign, Department of Molecular Biology, Aarhus
University, Gustav Wieds Vej 10 C, 8000, Aarhus C, Denmark.
The bacterial strain Flavobacterium sp. 4214 isolated from Greenland
was found to express beta-galactosidase (EC 3.2.1.23) at temperatures
below 25 degrees C. A chromosomal library of Flavobacterium sp. 4214
was constructed in Escherichia coli, and the gene gal4214-1 encoding a
beta-galactosidase of 1,046 amino acids (114.3 kDa) belonging to
glycosyl hydrolase family 2 was isolated. This was the only gene
encoding beta-galactosidase activity that was identified in the
chromosomal library. Expression levels in both Flavobacterium sp. 4214
and in initial recombinant E. coli strains were insufficient for
biochemical characterization. However, a combination of T7 promoter
expression and introduction of an E. coli host that complemented rare
transfer RNA genes yielded 15 mg of beta-galactosidase per liter of
culture. Gal4214-1-His protein was found to be active in monomeric
conformation. The protein was secreted from the cytoplasm, probably
through an N-terminal signaling sequence. The Gal4214-1-His protein was
found to have optimum activity at a temperature of 42 degrees C, but
with short-term stability at temperatures above 25 degrees C.
PMID: 16193277 [PubMed - as supplied by publisher]
9: J Biosci. 2005 Sep;30(4):469-74.
Structural organization of the transfer RNA operon I of Vibrio
cholerae: Differences between classical and El Tor strains.
Ghatak A, Majumdar A, Ghosh RK.
Molecular Biology Laboratory, Indian Institute of Chemical Biology, 4,
Raja SC Mullick Road,Jadavpur, Kolkata 700 032, India.
Nine major transfer RNA (tRNA) gene clusters were analysed in various
Vibrio cholerae strains. Of these, only the tRNA operon I was found to
differ significantly in V. cholerae classical (sixth pandemic) and El
Tor (seventh pandemic) strains. Amongst the sixteen tRNA genes
contained in this operon, genes for tRNA Gln3 (CAA) and tRNA Leu6 (CUA)
were absent in classical strains as compared to El Tor strains. The
observation strongly supported the view that the above two pandemic
strains constitute two different clones.
PMID: 16184008 [PubMed - in process]
10: Gene. 2005 Nov 7;360(2):92-102. Epub 2005 Sep 23.
The mitochondrial genome sequence of the scorpion Centruroides limpidus
(Karsch 1879) (Chelicerata; Arachnida).
Davila S, Pinero D, Bustos P, Cevallos MA, Davila G.
Programa de Genomica Evolutiva, Centro de Ciencias Genomicas - UNAM,
Avenida Universidad, s/n, Chamilpa, Cuernavaca 62210, Apartado Postal
565-A, Morelos, Mexico.
The mitochondrial genome of the scorpion Centruroides limpidus
(Chelicerata; Arachnida) has been completely sequenced and is 14519 bp
long. The genome contains 13 protein-encoding genes, two ribosomal RNA
genes, 21 transfer RNA genes and a large non-coding region related to
the control region. The overall A+T composition is the lowest among the
complete mitochondrial sequences published within the Chelicerata
subphylum. Gene order and gene content differ slightly from that of
Limulus polyphemus (Chelicerata: Xiphosura): i.e., the lack of the trnD
gene, and the translocation-inversion of the trnI gene. Preliminary
phylogenetic analysis of some Chelicerata shows that scorpions (C.
limpidus and Mesobuthus gibbosus) make a tight cluster with the spiders
(Arachnida; Araneae). Our analysis does not support that Scorpiones
order is the sister group to all Arachnida Class, since it is closer to
Araneae than to Acari orders.
PMID: 16183216 [PubMed - in process]
11: Nucleic Acids Res. 2005 Sep 15;33(16):5291-6. Print 2005.
Deacylated tRNA is released from the E site upon A site occupation but
before GTP is hydrolyzed by EF-Tu.
Dinos G, Kalpaxis DL, Wilson DN, Nierhaus KH.
Max-Planck-Institut fur Molekulare Genetik AG Ribosomen, Ihnestrasse
73, D-14195
Berlin, Germany.
The presence or absence of deacylated tRNA at the E site sharply
influences the activation energy required for binding of a ternary
complex to the ribosomal A site indicating the different conformations
that the E-tRNA imparts on the ribosome. Here we address two questions:
(i) whether or not peptidyltransferase--the essential catalytic
activity of the large ribosomal subunit--also depends on the occupancy
state of the E site and (ii) at what stage the E-tRNA is released
during an elongation cycle. Kinetics of the puromycin reaction on
various functional states of the ribosome indicate that the A-site
substrate of the peptidyltransferase center, puromycin, requires the
same activation energy for peptide-bond formation under all conditions
tested. We further demonstrate that deacylated tRNA is released from
the E site by binding a ternary complex aminoacyl-tRNA*EF-Tu*GDPNP to
the A site. This observation indicates that the E-tRNA is released
after the decoding step but before both GTP hydrolysis by EF-Tu and
accommodation of the A-tRNA. Collectively these results reveal that the
reciprocal linkage between the E and A sites affects the decoding
center on the 30S subunit, but does not influence the rate of peptidebond formation at the active center of the 50S subunit.
PMID: 16166657 [PubMed - indexed for MEDLINE]
12: Proc Natl Acad Sci U S A. 2005 Sep 20;102(38):13392-7. Epub 2005
Sep 12.
Crystal structure of a bacterial ribonuclease P RNA.
Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace
NR.
Department of Molecular, Cellular, and Developmental Biology,
University of Colorado, Boulder, CO 80309, USA.
The x-ray crystal structure of a 417-nt ribonuclease P RNA from
Bacillus stearothermophilus was solved to 3.3-A resolution. This RNA
enzyme is constructed from a number of coaxially stacked helical
domains joined together by local and long-range interactions. These
helical domains are arranged to form a remarkably flat surface, which
is implicated by a wealth of biochemical data in the binding and
cleavage of the precursors of transfer RNA substrate. Previous
photoaffinity crosslinking data are used to position the substrate on
the crystal structure and to identify the chemically active site of the
ribozyme. This site is located in a highly conserved core structure
formed by intricately interlaced long-range interactions between
interhelical sequences.
PMID: 16157868 [PubMed - in process]
13: Nucleic Acids Res. 2005 Sep 9;33(16):5120-32. Print 2005.
RNase P: role of distinct protein cofactors in tRNA substrate
recognition and RNA-based catalysis.
Sharin E, Schein A, Mann H, Ben-Asouli Y, Jarrous N.
Department of Molecular Biology, The Hebrew University-Hadassah Medical
School, Jerusalem 91120, Israel.
The Escherichia coli ribonuclease P (RNase P) has a protein component,
termed C5, which acts as a cofactor for the catalytic M1 RNA subunit
that processes the 5' leader sequence of precursor tRNA. Rpp29, a
conserved protein subunit of human RNase P, can substitute for C5
protein in reconstitution assays of M1 RNA activity. To better
understand the role of the former protein, we compare the mode of
action of Rpp29 to that of the C5 protein in activation of M1 RNA.
Enzyme kinetic analyses reveal that complexes of M1 RNA-Rpp29 and M1
RNA-C5 exhibit comparable binding affinities to precursor tRNA but
different catalytic efficiencies. High concentrations of substrate
impede the activity of the former complex. Rpp29 itself exhibits high
affinity in substrate binding, which seems to reduce the catalytic
efficiency of the reconstituted ribonucleoprotein. Rpp29
has a conserved C-terminal domain with an Sm-like fold that mediates
interaction with M1 RNA and precursor tRNA and can activate M1 RNA. The
results suggest that distinct protein folds in two unrelated protein
cofactors can facilitate transition from RNA- to ribonucleoproteinbased catalysis by RNase P.
PMID: 16155184 [PubMed - indexed for MEDLINE]
14: Gene. 2005 Nov 7;360(2):120-30. Epub 2005 Sep 8.
Congruence of evidence for a Methanopyrus-proximal root of life based
on transfer RNA and aminoacyl-tRNA synthetase genes.
Xue H, Ng SK, Tong KL, Wong JT.
Department of Biochemistry and Applied Genomics Laboratory, Hong Kong
University of Science and Technology, Hong Kong, PR China.
Among 60 organisms, the intraspecies genetic distances between tRNAs
cognate for different amino acids, between the initiator and elongator
tRNAs for Met, and between potentially paralogous pairs of aminoacyltRNA synthetases are found to be at a minimum within the Methanopyrus
kandleri genome. These results indicate an exact congruence between the
evidence from tRNA and aminoacyl-tRNA synthetase genes locating the
root of life closest to this organism.
PMID: 16153784 [PubMed - in process]
15: Biochemistry. 2005 Sep 13;44(36):12057-65.
Biochemical and Structural Studies of A-to-I Editing by tRNA:A34
Deaminases at the Wobble Position of Transfer RNA(,).
Elias Y, Huang RH.
Department of Biochemistry, School of Molecular and Cellular Biology,
University of Illinois, 600 South Mathews Avenue, Urbana, Illinois
61801.
Initial RNA transcription produces several tRNAs (one in prokaryotes
and plant chloroplasts and seven or eight in eukaryotes) that contain
an adenosine (A) at the wobble position (position 34). However, in all
cases, adenosine at position 34 is post-transcriptionally converted to
inosine (I), producing mature tRNAs without adenosine at the wobble
position. The enzymes responsible for this A-to-I conversion in tRNA
are tadA (acting as a homodimer) in prokaryotes and the heterodimeric
ADAT2-ADAT3 complex in eukaryotes. The genes encoding these proteins
are essential for cell viability, illustrating the biological
importance of A-to-I editing at the wobble position of tRNA. In this
study, recombinant tadA proteins from Escherichia coli, Agrobacterium
tumefaciens, and Aquifex aeolicus, as well as the ADAT2-ADAT3 proteins
from Saccharomyces cerevisiae, were overexpressed in E. coli and
purified to homogeneity by chromatography. Crystallization of a
proteolytically cleaved A. tumefaciens tadA (missing the last eight
amino acids at the C-terminus) produced high-quality crystals, and the
structure was determined at 1.6 A resolution. In addition, enzymatic
assays of the wild-type proteins as well as several mutants were
carried out using both the full-length E. coli tRNA(arg2) and the
truncated anticodon stem-loop motif as substrates. Our biochemical and
structural studies, in combination with sequence and structural
comparisons with other deaminases, allow us to propose a model of tadAtRNA interaction that explains the molecular basis of tRNA recognition
by tadA. In particular, a conserved FFxxxR motif at the C-terminus,
which is unique to tadA, has been identified, and its critical
role in tRNA substrate recognition is proposed. Furthermore, the
structural study of prokaryotic tadA presented here also sheds light on
tRNA substrate recognition and the possible evolutionary origin of the
eukaryotic ADAT2-ADAT3 heterodimer.
PMID: 16142903 [PubMed - in process]
16: Chembiochem. 2005 Oct 7;6(10):1805-1816.
A New Strategy for the Synthesis of Dinucleotides Loaded with
Glycosylated Amino Acids-Investigations on in vitro Non-natural Amino
Acid Mutagenesis for Glycoprotein Synthesis.
Rohrig CH, Retz OA, Hareng L, Hartung T, Schmidt RR.
Department of Chemistry, University of Konstanz, Fach M 725, 78457
Konstanz, Germany, Fax: (+49) 7531-883135.
The in vitro non-natural amino acid mutagenesis method provides the
opportunity to introduce non-natural amino acids site-specifically into
proteins. To this end, a chemically synthesised aminoacylated
dinucleotide is enzymatically ligated to a truncated suppressor
transfer RNA. The loaded suppressor tRNA is then used in translation
reactions to read an internal stop codon. Here we report an advanced
and general strategy for the synthesis of the aminoacyl dinucleotide.
The protecting group pattern developed for the dinucleotide facilitates
highly efficient aminoacylation, followed by one-step global
deprotection. The strategy was applied to the synthesis of
dinucleotides loaded with 2-acetamido-2-deoxy-glycosylated amino acids,
including N- and O-beta-glycosides and O- and C-alpha-glycosides of
amino acids, thus enabling the extension of in vitro non-natural amino
acid mutagenesis towards the synthesis of natural glycoproteins of high
biological interest. We demonstrate the incorporation of the
glycosylamino acids-although with low suppression efficiency-into the
human interleukin granulocyte-colony stimulating factor (hG-CSF), as
verified by the ELISA technique.
PMID: 16142818 [PubMed - as supplied by publisher]
17: Science. 2005 Sep 2;309(5740):1534-9.
Inositol hexakisphosphate is bound in the ADAR2 core and required for
RNA editing.
Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL.
Department of Biochemistry, University of Utah, Salt Lake City, UT
84132, USA.
We report the crystal structure of the catalytic domain of human ADAR2,
an RNA editing enzyme, at 1.7 angstrom resolution. The structure
reveals a zinc ion in the active site and suggests how the substrate
adenosine is recognized. Unexpectedly, inositol hexakisphosphate (IP6)
is buried within the enzyme core, contributing to the protein fold.
Although there are no reports that adenosine deaminases that act on RNA
(ADARs) require a cofactor, we show that IP6 is required for activity.
Amino acids that coordinate IP6 in the crystal structure are conserved
in some adenosine deaminases that act on transfer RNA (tRNA) (ADATs),
related enzymes that edit tRNA. Indeed, IP6 is also essential for in
vivo and in vitro deamination of adenosine 37 of tRNAala by ADAT1.
PMID: 16141067 [PubMed - indexed for MEDLINE]
18: Science. 2005 Sep 2;309(5740):1508-14.
RNA structure: reading the ribosome.
Noller HF.
Center for Molecular Biology of RNA, Department of Molecular, Cell, and
Developmental Biology, Sinsheimer Laboratories, University of
California, Santa Cruz, Santa Cruz, CA 95064, USA.
The crystal structures of the ribosome and its subunits have increased
the amount of information about RNA structure by about two orders of
magnitude. This is leading to an understanding of the principles of RNA
folding and of the molecular interactions that underlie the functional
capabilities of the ribosome and other RNA systems. Nearly all of the
possible types of RNA tertiary interactions have been found in
ribosomal RNA. One of these, an abundant tertiary structural motif
called the A-minor interaction, has been shown to participate in both
aminoacyl-transfer RNA selection and in peptidyl transferase; it may
also play an important role in the structural dynamics of the ribosome.
Publication Types:
Review
Review, Tutorial
PMID: 16141058 [PubMed - indexed for MEDLINE]
19: Mol Cell. 2005 Sep 2;19(5):707-16.
Barrier function at HMR.
Oki M, Kamakaka RT.
Unit on Chromatin and Transcription, National Institute of Child Health
and Human Development, National Institutes of Health, Bethesda,
Maryland 20892, USA.
The silenced HMR domain is restricted from spreading by barrier
elements, and the right barrier is a unique t-RNA(THR) gene. We show
that sequences immediately flanking the silenced domain were enriched
in acetylated, but not methylated, histones, whereas the barrier
element was associated with a nucleosome-free region. Surprisingly, the
SAGA acetyltransferase resided across the entire region. We further
demonstrate that a mutation in the barrier eliminated the nucleosomefree gap but only subtly altered the distribution of SAGA.
Interestingly, neither reformation of the nucleosome nor mutations in
chromatin-modifying enzymes alone led to an unrestricted spread of
silenced chromatin. Double mutations in the t-RNA barrier and these
complexes, on the other hand, led to a significant spread of Sir
proteins. These results suggest two overlapping mechanisms function to
restrict the spread of silencing: one of which involves a DNA binding
element, whereas the other mechanism involves specific chromatinmodifying activities.
PMID: 16137626 [PubMed - indexed for MEDLINE]
20: Nat Struct Mol Biol. 2005 Sep;12(9):788-93. Epub 2005 Aug 21.
Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding.
Olejniczak M, Dale T, Fahlman RP, Uhlenbeck OC.
Department of Biochemistry, Molecular Biology and Cell Biology,
Northwestern University, 2145 Sheridan Road, Evanston, Illinois 60208,
USA.
The binding of seven tRNA anticodons to their complementary codons on
Escherichia coli ribosomes was substantially impaired, as compared with
the binding of their natural tRNAs, when they were transplanted into
tRNA(2)(Ala). An analysis of chimeras composed of tRNA(2)(Ala) and
various amounts of either tRNA(3)(Gly) or tRNA(2)(Arg) indicates that
the presence of the parental 32-38 nucleotide pair is sufficient to
restore ribosome binding of the transplanted anticodons. Furthermore,
mutagenesis of tRNA(2)(Ala) showed that its highly conserved A32-U38
pair serves to weaken ribosome affinity. We propose that this negative
binding determinant is used to offset the very tight codon-anticodon
interaction of tRNA(2)(Ala). This suggests that each tRNA sequence has
coevolved with its anticodon to tune ribosome affinity to a value that
is the same for all tRNAs.
PMID: 16116437 [PubMed - indexed for MEDLINE]
21: Proc Natl Acad Sci U S A. 2005 Aug 30;102(35):12395-400. Epub 2005
Aug 22.
Mechanism of peptide bond synthesis on the ribosome.
Trobro S, Aqvist J.
Department of Cell and Molecular Biology, Uppsala Biomedical Center,
Uppsala University, Box 596, SE-751 24 Uppsala, Sweden.
With the emergence of atomic-resolution crystal structures of bacterial
ribosomal subunits, major advances in eliciting structure-function
relationships of the translation process are underway. Nevertheless,
the detailed mechanism of peptide bond synthesis that occurs on the
large ribosomal subunit remains unknown. Separate x-ray structures of
aminoacyl-tRNA and peptidyl-tRNA analogues bound to the ribosomal Aand P-sites, however, allow for structural modeling of the active
complex in catalysis. Here, we combine available structural data to
construct such a model of the peptidyl transfer reaction center with
bound substrates. Molecular dynamics and free energy perturbation
simulations then are used in combination with an empirical valence bond
description of the reaction energy surface to examine possible
catalytic mechanisms. Already, simulations of the reactant and
tetrahedral intermediate states reveal a stable, preorganized
H-bond network poised for catalysis. The most favorable mechanism is
found not to involve any general acid-base catalysis by ribosomal
groups but an intra-reactant proton shuttling via the P-site adenine
O2' oxygen, which follows the attack of the A-site alpha-amino group on
the P-site ester. The calculated rate enhancement for this mechanism is
approximately 10(5), and the catalytic effect is found to be entirely
of entropic origin, in accordance with recent experimental data, and is
associated with the reduction of solvent reorganization energy rather
than with substrate alignment or proximity. This mechanism also
explains the inability of 2'-deoxyadenine P-site substrates to
promote peptidyl transfer. The observed H-bond network suggests an
important structural role of several universally conserved rRNA
residues.
PMID: 16116099 [PubMed - indexed for MEDLINE]
22: Nature. 2005 Sep 22;437(7058):584-7. Epub 2005 Aug 21.
Crystal structure of the RNA component of bacterial ribonuclease P.
Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A.
Department of Biochemistry, Molecular Biology and Cell Biology,
Northwestern University, Evanston, Illinois 60208, USA.
Transfer RNA (tRNA) is produced as a precursor molecule that needs to
be processed at its 3' and 5' ends. Ribonuclease P is the sole
endonuclease responsible for processing the 5' end of tRNA by cleaving
the precursor and leading to tRNA maturation. It was one of the first
catalytic RNA molecules identified and consists of a single RNA
component in all organisms and only one protein component in bacteria.
It is a true multi-turnover ribozyme and one of only two ribozymes (the
other being the ribosome) that are conserved in all kingdoms of life.
Here we show the crystal structure at 3.85 A resolution of the RNA
component of Thermotoga maritima ribonuclease P. The entire RNA
catalytic component is revealed, as well as the arrangement of the two
structural domains. The structure shows the general architecture of the
RNA molecule, the inter- and intra-domain interactions, the location of
the universally conserved regions, the regions involved in pre-tRNA
recognition and the location of the active site. A model with bound
tRNA is in agreement with all existing data and suggests the general
basis for RNA-RNA recognition by this ribozyme.
PMID: 16113684 [PubMed - indexed for MEDLINE]
23: Nucleic Acids Res. 2005 Aug 19;33(15):4683-91. Print 2005.
A unique tRNA recognition mechanism of Caenorhabditis elegans
mitochondrial
EF-Tu2.
Suematsu T, Sato A, Sakurai M, Watanabe K, Ohtsuki T.
Department of Integrated Bioscience, Graduate School of Frontier
Science, The University of Tokyo 5-1-5 Kashiwanoha, Kashiwa, 277-8562,
Japan.
Nematode mitochondria expresses two types of extremely truncated tRNAs
that are specifically recognized by two distinct elongation factor Tu
(EF-Tu) species named EF-Tu1 and EF-Tu2. This is unlike the canonical
EF-Tu molecule that participates in the standard protein biosynthesis
systems, which basically recognizes all elongator tRNAs. EF-Tu2
specifically recognizes Ser-tRNA(Ser) that lacks a D arm but has a
short T arm. Our previous study led us to speculate the lack of the D
arm may be essential for the tRNA recognition of EF-Tu2. However, here,
we showed that the EF-Tu2 can bind to D arm-bearing Ser-tRNAs, in which
the D-T arm interaction was weakened by the mutations. The
ethylnitrosourea-modification interference assay showed that EF-Tu2 is
unique, in that it interacts with the phosphate groups on the T stem on
the side that is opposite to where canonical EF-Tu binds. The
hydrolysis protection assay using several EF-Tu2 mutants then strongly
suggests that seven C-terminal amino acid residues of EF-Tu2 are
essential for its aminoacyl-tRNA-binding activity. Our results indicate
that the formation of the nematode mitochondrial (mt)EFTu2/GTP/aminoacyl-tRNA ternary complex is probably supported by a
unique interaction between the C-terminal extension of EF-Tu2 and the
tRNA.
PMID: 16113240 [PubMed - indexed for MEDLINE]
*****THROWN OUT – NO ABSTRACT*****
24: Nippon Rinsho. 2005 Jul;63 Suppl 7:505-7.
[Anti-Jo-l antibody (anti-histidyl transfer RNA synthetase antibody)]
[Article in Japanese]
Katsuki Y, Hirakata M.
Department of Internal Medicine, Keio University School of Medicine.
PMID: 16111315 [PubMed - in process]
***** END THROW OUT *****
25: Mol Ecol. 2005 Sep;14(10):3077-94.
The influence of Pleistocene glacial refugia on tawny owl genetic
diversity and phylogeography in western Europe.
Brito PH.
American Museum of Natural History, Ornithology Department, Central
Park West at 79th Street, New York, NY 10024, USA. pbrito@amnh.org
The glacial refugia hypothesis indicates that during the height of the
Pleistocene glaciations the temperate species that are today widespread
in western Europe must have survived in small and climatically
favourable areas located in the southern peninsulas of Iberia, Italy
and Balkans. One such species is the tawny owl, a relatively sedentary,
nonmigratory bird presently distributed throughout Europe. It is a
tree-nesting species closely associated with deciduous and mixed
coniferous woodlands. In this study I used control region mtDNA
sequences from 187 individuals distributed among 14 populations to
determine whether current genetic patterns in tawny owl populations
were consistent with postglacial expansion from peninsular refugia.
European, North African and Asian tawny owls were found to represent
three distinct lineages, where North Africa is the sister clade to all
European owls. Within Europe, I found three well-supported clades that
correspond to each of the three allopatric refugia. Expansion patterns
indicate that owls from the Balkan refugium repopulated most of
northern Europe, while expansion out of Iberia and Italy had only
regional effects leading to admixture in France. Estimates of
population divergence times between refugia populations are roughly
similar, but one order of magnitude smaller between Greece and northern
Europe. Based on a wide range of mutation rates and generation times,
divergence between refugia appears to date to the Pleistocene.
PMID: 16101775 [PubMed - indexed for MEDLINE]
26: Mol Ecol. 2005 Sep;14(10):2991-3004.
Ice age cloning--comparison of the Quaternary evolutionary histories of
sexual and clonal forms of spiny loaches (Cobitis; Teleostei) using the
analysis of mitochondrial DNA variation.
Janko K, Culling MA, Rab P, Kotlik P.
Laboratory of Fish Genetics, Institute of Animal Physiology and
Genetics, Academy of Sciences of the Czech Republic, Rumburska 89,
27721 Libechov, Czech Republic. janko@iapg.cas.cz
Recent advances in population history reconstruction offered a powerful
tool for comparisons of the abilities of sexual and clonal forms to
respond to Quaternary climatic oscillations, ultimately leading to
inferences about the advantages and disadvantages of a given mode of
reproduction. We reconstructed the Quaternary historical biogeography
of the sexual parental species and clonal hybrid lineages within the
Europe-wide hybrid complex of Cobitis spiny loaches. Cobitis
elongatoides and Cobitis taenia recolonizing Europe from separated
refuges met in central Europe and the Pontic region giving rise to
hybrid lineages during the Holocene. Cobitis elongatoides due to its
long-term reproductive contact with the remaining parental species of
the complex--C. tanaitica and C. spec.--gave rise to two clonal hybrid
lineages probably during the last interglacial or even earlier, which
survived the Wurmian glaciation with C. elongatoides. These lineages
followed C. elongatoides postglacial expansion and probably decreased
its dispersal rate. Our data indicate the frequent origins of
asexuality irrespective of the parental populations involved and the
comparable dispersal potential of diploid and triploid lineages.
PMID: 16101769 [PubMed - indexed for MEDLINE]
27: Biochemistry. 2005 Aug 23;44(33):11254-61.
Predicting the binding affinities of misacylated tRNAs for Thermus
thermophilus EF-Tu.GTP.
Asahara H, Uhlenbeck OC.
Department of Biochemistry, Molecular Biology, and Cell Biology,
Northwestern University, 2205 Tech Drive, Hogan 2-100, Evanston,
Illinois 60208, USA.
The free energies for the binding of 20 different unmodified
Escherichia coli elongator aminoacyl-tRNAs to Thermus thermophilus
elongation factor Tu (EF-Tu) were determined. When combined with the
binding free energies for the same tRNA bodies misacylated with either
valine or phenylalanine determined previously [Asahara, H., and
Uhlenbeck, O. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99,
3499-3504], these data permit the calculation of the contribution of
each esterified amino acid to the total free energy of binding of the
complex. The two data sets can also be used to calculate the free
energy of binding of EF-Tu to any misacylated E. coli tRNA, and the
values agree well with previously published experimental values. In
addition, a survey of active misacylated suppressor tRNAs suggests that
a minimal threshold of binding free energy for EF-Tu is required for
suppression to occur.
PMID: 16101309 [PubMed - indexed for MEDLINE]
28: Cytogenet Genome Res. 2005;110(1-4):288-98.
Transfer RNA gene-targeted integration: an adaptation of
retrotransposable elements to survive in the compact Dictyostelium
discoideum genome.
Winckler T, Szafranski K, Glockner G.
Institut fur Pharmazeutische Biologie, Universitat Frankfurt am Main
(Biozentrum), Frankfurt, Germany. Winckler@em-uni-frankfurt.de
Almost every organism carries along a multitude of molecular parasites
known as transposable elements (TEs). TEs influence their host genomes
in many ways by expanding genome size and complexity, rearranging
genomic DNA, mutagenizing host genes, and altering transcription levels
of nearby genes. The eukaryotic microorganism Dictyostelium discoideum
is attractive for the study of fundamental biological phenomena such as
intercellular communication, formation of multicellularity, cell
differentiation, and morphogenesis. D. discoideum has a highly
compacted, haploid genome with less than 1 kb of genomic DNA separating
coding regions. Nevertheless, the D. discoideum genome is loaded with
10% of TEs that managed to settle and survive in this inhospitable
environment. In depth analysis of D. discoideum genome project data has
provided intriguing insights into the evolutionary challenges that
mobile elements face when they invade compact genomes. Two different
mechanisms are used by D. discoideum TEs to avoid disruption of host
genes upon retrotransposition. Several TEs have invented the specific
targeting of tRNA gene-flanking regions as a means to avoid integration
into coding regions. These elements have been dispersed on all
chromosomes, closely following the distribution of tRNA genes. By
contrast, TEs that lack bona fide integration specificities show a
strong bias to nested integration, thus forming large TE clusters at
certain chromosomal loci that are hardly resolved by bioinformatics
approaches. We summarize our current view of D. discoideum TEs and
present new data from the analysis of the complete sequences
of D. discoideum chromosomes 1 and 2, which comprise more than one
third of the total genome.
PMID: 16093681 [PubMed - in process]
29: BMC Genomics. 2005 Aug 9;6:105.
The complete mitochondrial genome of the stomatopod crustacean Squilla
mantis.
Cook CE.
Department and Museum of Zoology, University of Cambridge, Downing
Street, Cambridge, CB2 3EJ, UK. cecook@alum.exeter.edu
BACKGROUND: Animal mitochondrial genomes are physically separate from
the much larger nuclear genomes and have proven useful both for
phylogenetic studies and for understanding genome evolution. Within the
phylum Arthropoda the subphylum Crustacea includes over 50,000 named
species with immense variation in body plans and habitats, yet only 23
complete mitochondrial genomes are available from this subphylum.
RESULTS: I describe here the complete mitochondrial genome
of the crustacean Squilla mantis (Crustacea: Malacostraca:
Stomatopoda). This 15994-nucleotide genome, the first described from a
hoplocarid, contains the standard complement of 13 protein-coding
genes, 22 transfer RNA genes, two ribosomal RNA genes, and a non-coding
AT-rich region that is found in most other metazoans. The gene order is
identical to that considered ancestral for hexapods and crustaceans.
The 70% AT base composition is within the range described for other
arthropods. A single unusual feature of the genome is a 230 nucleotide
non-coding region between a serine transfer RNA and the nad1 gene,
which has no apparent function. I also compare gene order, nucleotide
composition, and codon usage of the S. mantis genome and eight other
malacostracan crustaceans. A translocation of the histidine transfer
RNA gene is shared by three taxa in the order Decapoda, infraorder
Brachyura; Callinectes sapidus, Portunus trituberculatus and
Pseudocarcinus gigas. This translocation may be diagnostic for the
Brachyura. For all nine taxa nucleotide composition is biased towards
AT-richness, as expected for arthropods, and is within the range
reported for other arthropods. Codon usage is biased, and much of this
bias is probably due to the skew in nucleotide composition towards ATrichness. CONCLUSION: The mitochondrial genome of Squilla mantis
contains one unusual feature, a 230 base pair non-coding region has so
far not been described in any other malacostracan. Comparisons with
other Malacostraca show that all nine genomes, like most other
mitochondrial genomes, share a bias toward AT-richness and a related
bias in codon usage. The nine malacostracans included in this analysis
are not representative of the diversity of the class Malacostraca, and
additional malacostracan sequences would surely reveal other unusual
genomic features that could be useful in understanding mitochondrial
evolution in this taxon.
PMID: 16091132 [PubMed - in process]
30: Nucleic Acids Res. 2005 Aug 5;33(14):4433-42. Print 2005.
The Rhodomonas salina mitochondrial genome: bacteria-like operons,
compact gene arrangement and complex repeat region.
Hauth AM, Maier UG, Lang BF, Burger G.
Departement de Biochimie, Robert Cedergren Research Center for
Bioinformatics and Genomics, Canadian Institute for Advanced Research,
Universite de Montreal 2900 Boulevard Edouard-Montpetit, Montreal,
Quebec, Canada H3T 1J4. amy.hauth@umontreal.ca
To gain insight into the mitochondrial genome structure and gene
content of a putatively ancestral group of eukaryotes, the
cryptophytes, we sequenced the complete mitochondrial DNA of Rhodomonas
salina. The 48 063 bp circular-mapping molecule codes for 2 rRNAs, 27
tRNAs and 40 proteins including 23 components of oxidative
phosphorylation, 15 ribosomal proteins and two subunits of tat
translocase. One potential protein (ORF161) is without assigned
function. Only two introns occur in the genome; both are present within
cox1 belong to group II and contain RT open reading frames. Primitive
genome features include bacteria-like rRNAs and tRNAs, ribosomal
protein genes organized in large clusters resembling bacterial operons
and the presence of the otherwise rare genes such as rps1 and tatA. The
highly compact gene organization contrasts with the presence of a 4.7
kb long, repeat-containing intergenic region. Repeat motifs
approximately 40-700 bp long occur up to 31 times, forming a complex
repeat structure. Tandem repeats are the major arrangement but the
region also includes a large, approximately 3 kb, inverted repeat and
several potentially stable approximately 40-80 bp long hairpin
structures. We provide evidence that the large repeat region is
involved in replication and transcription initiation, predict a
promoter motif that occurs in three locations and discuss two likely
scenarios of how this highly structured repeat region might have
evolved.
PMID: 16085754 [PubMed - indexed for MEDLINE]
31: J Mol Biol. 2005 Sep 9;352(1):22-7.
RNase E cleavage in the 5' leader of a tRNA precursor.
Soderbom F, Svard SG, Kirsebom LA.
Department of Molecular Biology, Swedish University of Agricultural
Sciences, Box 590, Biomedical Centre, SE-751 24 Uppsala, Sweden.
In this study, we have used various tRNA(Tyr)Su3 precursor (pSu3)
derivatives that are processed less efficiently by RNase P to
investigate if the 5' leader is a target for RNase E. We present data
that suggest that RNase E cleaves the 5' leader of pSu3 both in vivo
and in vitro. The site of cleavage in the 5' leader corresponds to the
cleavage site for a previously identified endonuclease activity
referred to as RNase P2/O. Thus, our findings suggest that RNase P2/O
and RNase E activities are of the same origin. These data are in
keeping with the suggestion that the structure of the 5' leader
influences tRNA expression by affecting tRNA processing and indicate
the involvement of RNase E in the regulation of cellular tRNA levels.
PMID: 16081101 [PubMed - indexed for MEDLINE]
32: Am J Med Genet A. 2005 Aug 30;137(2):170-5.
A mitochondrial tRNA aspartate mutation causing isolated mitochondrial
myopathy.
Seneca S, Goemans N, Van Coster R, Givron P, Reybrouck T, Sciot R,
Meulemans A, Smet J, Van Hove JL.
Center of Medical Genetics, Free University Brussels, Brussels,
Belgium.
Several mutations in mitochondrial transfer RNA (tRNA) genes can cause
mitochondrial myopathy. We describe a young girl who presented with
pronounced exercise intolerance. The anaerobic threshold and the
maximal oxygen consumption were decreased. She had decreased complex I
and IV enzyme activity and ragged red fibers on muscle biopsy. An A to
G transition at nucleotide position 7526 in tRNA Aspartate (tRNA(Asp))
gene was heteroplasmic in several of the patient's tissues. We were
unable to detect the mutation in muscle tissue from the patient's
mother. This case adds a new genetic etiology for mitochondrial
myopathy. It also illustrates for patients with combined deficiency of
the complex I and IV enzyme activity the value of sequencing in the
affected tissue muscle, and not only in blood, all mitochondrial tRNA
genes including those not commonly affected, such as in this case mt
tRNA(Asp). (c) 2005 Wiley-Liss, Inc.
Publication Types:
Case Reports
PMID: 16059939 [PubMed - indexed for MEDLINE]
KEYWORD SEARCH: “small nuclear RNA”
1: Br J Dermatol. 2005 Nov;153(5):981-6.
The role of CD4 and CD8 cytotoxic T lymphocytes in the formation of
viral vesicles.
Morizane S, Suzuki D, Tsuji K, Oono T, Iwatsuki K.
Department of Dermatology, Okayama University Graduate School of
Medicine and Dentistry, 2-5-1, Shikata-cho, Okayama, 700-8558, Japan.
Background Herpetic vesicles caused by herpes simplex virus and
varicella zoster virus, and hydroa vacciniforme (HV) are characterized
by umbilicated vesicule formation. Objectives To understand the
histogenesis of umbilicated vesicles in herpetic vesicles and HV, we
demonstrated the presence of the virus-associated molecules in the
lesions, and the pathogenic role of cytotoxic T-lymphocyte (CTL) immune
responses. Methods Phenotyping of infiltrating cells was carried
out in biopsy specimens from herpes simplex, varicella, herpes zoster
and HV, and compared with nonviral contact dermatitis. Viral antigens
and Epstein-Barr virus-encoded small nuclear RNA (EBER) were detected
by immunostaining and by in situ hybridization, respectively.
Infiltrating CTLs expressing granzyme B and granulysin were determined
by double immunostaining using confocal laser scanning microscopy.
Results In all herpetic vesicles, the corresponding viral
antigens were observed in the cytopathic keratinocytes, and
infiltration of lymphoid cells was present in the upper dermis and
around the vessels. In all HV lesions studied, EBER+ T cells made up 510% of the dermal infiltrates and the dermal infiltrates contained
almost no CD56 cells. CTLs expressing granzyme B and granulysin were
present in both herpetic and HV lesions, in which they made
up 10-30% of the total dermal infiltrates, whereas they comprised less
than 5% of the infiltrates of biopsy specimens from nonviral contact
dermatitis. Confocal laser microscopic examination demonstrated that
both CD4+ and CD8+ T cells expressed granzyme B and granulysin.
Conclusions CD4+ and/or CD8+ CTLs reactive to the virus-infected cells
might be responsible for the histogenesis of herpetic and HV lesions
characterized by umbilicated vesicles.
PMID: 16225610 [PubMed - in process]
2: Nucleic Acids Res. 2005 Sep 9;33(16):5112-9. Print 2005.
Extended base pair complementarity between U1 snRNA and the 5' splice
site does not inhibit splicing in higher eukaryotes, but rather
increases 5' splice site recognition.
Freund M, Hicks MJ, Konermann C, Otte M, Hertel KJ, Schaal H.
Institut fur Virologie, Heinrich-Heine-Universitat Dusseldorf,
Universitatsstr. 1, Geb. 22.21, D-40225 Dusseldorf, Germany.
Spliceosome formation is initiated by the recognition of the 5' splice
site through formation of an RNA duplex between the 5' splice site and
U1 snRNA. We have previously shown that RNA duplex formation between U1
snRNA and the 5' splice site can protect pre-mRNAs from degradation
prior to splicing. This initial RNA duplex must be disrupted to expose
the 5' splice site sequence for base pairing with U6 snRNA and to form
the active spliceosome. Here, we investigated whether
hyperstabilization of the U1 snRNA/5' splice site duplex
interferes with splicing efficiency in human cell lines or nuclear
extracts. Unlike observations in Saccharomyces cerevisiae, we
demonstrate that an extended U1 snRNA/5' splice site interaction does
not decrease splicing efficiency, but rather increases 5' splice site
recognition and exon inclusion. However, low complementarity of the 5'
splice site to U1 snRNA significantly increases exon skipping and RNA
degradation. Although the splicing mechanisms are conserved between
human and S.cerevisiae, these results demonstrate that distinct
differences exist in the activation of the spliceosome.
PMID: 16155183 [PubMed - indexed for MEDLINE]
3: J Cell Physiol. 2005 Dec;205(3):463-70.
siRNA depletion of 7SK snRNA induces apoptosis but does not affect
expression of the HIV-1 LTR or P-TEFb-dependent cellular genes.
Haaland RE, Herrmann CH, Rice AP.
Department of Molecular Virology and Microbiology, Baylor College of
Medicine, Houston, Texas.
P-TEFb is a general transcriptional elongation factor composed of Cdk9
and either cyclin T1, T2, or K. A substantial portion of P-TEFb is
associated with the 7SK small nuclear RNA (7SK) and the HEXIM1 or
HEXIM2 proteins; this complex has reduced kinase activity in vitro
relative to free P-TEFb. Here we report that 7SK and HEXIM1 levels are
induced in activated lymphocytes concomitantly with increased P-TEFb
activity and global transcription. We used siRNA-mediated depletion to
probe the function of 7SK in HeLa cells. Depletion of 7SK caused a
large reduction in the association of HEXIM1 with Cdk9 and cyclin T1,
and greatly reduced the amount of the cyclin T1 present in the
7SK/HEXIM1/P-TEFb complex. Similar to previous studies, siRNA-mediated
depletion of 7SK resulted in increased expression of several reporter
plasmids tested, including a plasmid lacking promoter elements.
However, in contrast to previous studies, which did not examine the
effects of 7SK depletion on endogenous gene expression, depletion of
7SK did not appear to affect the expression of the corresponding
endogenous genes. Moreover, 7SK depletion had no effect on expression
from the integrated HIV-1 provirus or the c-myc and MCL-1 genes, three
transcription units known to be highly dependent upon P-TEFb.
Importantly, depletion of 7SK was found to cause apoptosis by 72 h
post-transfection in HeLa cells. These results suggest that 7SK may
provide an essential cellular function whose relation to P-TEFb
function is unclear. (c) 2005 Wiley-Liss, Inc.
PMID: 16152622 [PubMed - in process]
4: Mol Cell Probes. 2005 Sep 5; [Epub ahead of print]
Nonisotopic detection of microRNA using digoxigenin labeled RNA probes.
Ramkissoon SH, Mainwaring LA, Sloand EM, Young NS, Kajigaya S.
Howard Hughes Medical Institute-National Institutes of Health Research
Scholars Program, Graduate School of Biomedical Sciences, University of
Medicine and Dentistry of New Jersey, Bethesda, MD, USA; Hematology
Branch, National Heart, Lung, and Blood Institute, National Institutes
of Health, 9000 Rockville Pike, Bethesda, MD 20892-1202, USA.
MicroRNAs (miRNAs) are an important class of endogenously derived,
small approximately 22 nucleotide noncoding regulatory RNAs that have
recently become implicated in development, cell regulation and cancers
of various tissues. Here we report a nonisotopic Northern analysis
method for miRNA detection using 3'-digoxigenin (DIG)-labeled RNA oligo
probes. Northern blot analysis was performed using miRNA or total RNA
fractions extracted from human leukemic cell lines, and blots were
hybridized with either (32)P- or DIG-labeled RNA probe for miR-181,
miR-155 or miR-16. A labeled probe for U6 small nuclear RNA served as
an internal control. The use of DIG-labeled RNA probes was equally
sensitive compared to (32)P-labeled probes in detecting miRNA
quantities as low as 50ng. The ability to use nonisotopic methods and
yet obtain sensitive and reliable results offers an advantage to
investigators who prefer to avoid isotopes.
PMID: 16146683 [PubMed - as supplied by publisher]
5: Mol Cell. 2005 Aug 19;19(4):523-34.
The bromodomain protein Brd4 is a positive regulatory component of PTEFb and stimulates RNA polymerase II-dependent transcription.
Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K.
Laboratory of Molecular Growth Regulation, National Institute of Child
Health and Human Development, National Institutes of Health, Bethesda,
Maryland 20892, USA.
Brd4 is a mammalian bromodomain protein that binds to acetylated
chromatin. Proteomic analysis revealed that Brd4 interacts with
cyclinT1 and Cdk9 that constitutes core positive transcription
elongation factor b (P-TEFb). Brd4 interacted with P-TEFb in the living
nucleus through its bromodomain. About half of P-TEFb was bound to the
inhibitory subunit and functionally inactive. Brd4 interacted with PTEFb that was free of the inhibitory subunit. An increase in Brd4
expression led to increased P-TEFb-dependent phosphorylation of RNA
polymerase II (RNAPII) CTD and stimulation of transcription from
promoters in vivo. Conversely, a reduction in Brd4 expression by siRNA
reduced CTD phosphorylation and transcription, revealing that Brd4 is a
positive regulatory component of P-TEFb. In chromatin
immunoprecipitation (ChIP) assays, the recruitment of P-TEFb to a
promoter was dependent on Brd4 and was enhanced by an increase in
chromatin acetylation. Together, P-TEFb alternately interacts with
Brd4 and the inhibitory subunit to maintain functional equilibrium in
the cell.
PMID: 16109376 [PubMed - indexed for MEDLINE]
6: J Biol Chem. 2005 Oct 14;280(41):34558-68. Epub 2005 Aug 17.
Elucidating the Role of H/ACA-like RNAs in trans-Splicing and rRNA
Processing via RNA Interference Silencing of the Trypanosoma brucei
CBF5 Pseudouridine Synthase.
Barth S, Hury A, Liang XH, Michaeli S.
Faculty of Life Sciences, Bar-Ilan University, Ramat-Gan 52900, Israel.
Most pseudouridinylation in eukaryotic rRNA and small nuclear RNAs is
guided by H/ACA small nucleolar RNAs. In this study, the Trypanosoma
brucei pseudouridine synthase, Cbf5p, a snoRNP protein, was identified
and silenced by RNAi. Depletion of this protein destabilized all small
nucleolar RNAs of the H/ACA-like family. Following silencing, defects
in rRNA processing, such as accumulation of precursors and inhibition
of cleavages to generate the mature rRNA, were observed. snR30, an
H/ACA RNA involved in rRNA maturation, was identified based on
prototypical conserved domains characteristic of this RNA in other
eukaryotes. The silencing of CBF5 also eliminated the spliced
leader-associated (SLA1) RNA that directs pseudouridylation on the
spliced leader RNA (SL RNA), which is the substrate for the transsplicing reaction. Surprisingly, the depletion of Cbf5p not only
eliminated the pseudouridine on the SL RNA but also abolished capping
at the fourth cap-4 nucleotide. As a result of defects in the SL RNA
and decreased modification on the U small nuclear RNA, trans-splicing
was inhibited at the first step of the reaction, providing evidence for
the essential role of H/ACA RNAs and the modifications they guide on
trans-splicing.
PMID: 16107339 [PubMed - in process]
7: Genes Dev. 2005 Aug 15;19(16):1894-904.
The Isy1p component of the NineTeen complex interacts with the ATPase
Prp16p to regulate the fidelity of pre-mRNA splicing.
Villa T, Guthrie C.
Department of Biochemistry and Biophysics, University of California,
San Francisco, 94143-2200, USA.
Prp16p is a DEAH-box ATPase that transiently associates with the
spliceosome to promote the structural transition required for the
second chemical step. Yeast strains carrying the cold-sensitive allele
prp16-302 stall the release of Prp16p at low temperatures, yet splice
precursors with aberrant branchpoints at increased frequency. To
identify new factors involved in the regulation of splicing fidelity,
we sought suppressors of the prp16-302 growth phenotype. Deletion of
the nonessential ISY1 gene (1) improves growth of prp16-302 strains,
(2) alleviates stalling, and (3) restores fidelity of branchpoint usage
to wild-type levels. Isy1p is a subunit of the NineTeen Complex
containing Prp19p, an essential E3 ubiquitin ligase homolog required
for splicing. Notably, Deltaisy1 PRP16 strains display reduced fidelity
of 3'-splice site selection. Consistent with a recent two-state model
of the spliceosome, our genetic and biochemical results suggest that
Isy1p acts together with U6 snRNA to promote a spliceosomal
conformation favorable for first-step chemistry. We propose that
deletion of ISY1 favors the premature release of Prp16p, thus promoting
second-step chemistry of precursors with inappropriate 3'-splice sites.
PMID: 16103217 [PubMed - indexed for MEDLINE]
8: Plant J. 2005 Aug;43(4):479-90.
Involvement of SRD2-mediated activation of snRNA transcription in the
control of cell proliferation competence in Arabidopsis.
Ohtani M, Sugiyama M.
Botanical Gardens, Graduate School of Science, The University of Tokyo,
3-7-1 Hakusan, Bunkyo-ku, Tokyo 112-0001, Japan.
The transcription machinery of small nuclear RNA (snRNA) genes has been
investigated extensively in human cell-free systems, but its
physiological function in vivo has not been addressed. This paper
demonstrates the physiological role of an activator of snRNA
transcription using a temperature-sensitive mutant of Arabidopsis
thaliana, srd2. Phenotypic characteristics of the srd2 mutant suggest
that the SRD2 gene participates in the control of competence in cell
proliferation. The SRD2 gene encodes a nuclear protein that shares
sequence similarity with the human SNAP50 protein, which is a subunit
of SNAPc and is required for snRNA transcription in vitro. Our
results, obtained from analysis of snRNA expression in the srd2 mutant,
indicate that the SRD2 protein functions in the upregulation of
transcription of snRNA genes, the promoters of which contain the
upstream sequence element, to elevate cell proliferation competence.
PMID: 16098103 [PubMed - in process]
9: Mol Cell Biol. 2005 Aug;25(16):7314-22.
Trypanosomal TBP functions with the multisubunit transcription factor
tSNAP to direct spliced-leader RNA gene expression.
Das A, Zhang Q, Palenchar JB, Chatterjee B, Cross GA, Bellofatto V.
Department of Microbiology and Molecular Genetics, University of
Medicine and Dentistry of New Jersey-New Jersey Medical School, Newark,
07103, USA.
Protein-coding genes of trypanosomes are mainly transcribed
polycistronically and cleaved into functional mRNAs in a process that
requires trans splicing of a capped 39-nucleotide RNA derived from a
short transcript, the spliced-leader (SL) RNA. SL RNA genes are
individually transcribed from the only identified trypanosome RNA
polymerase II promoter. We have purified and characterized a sequencespecific SL RNA promoter-binding complex, tSNAP(c), from the pathogenic
parasite Trypanosoma brucei, which induces robust transcriptional
activity within the SL RNA gene. Two tSNAP(c) subunits resemble
essential components of the metazoan transcription factor SNAP(c),
which directs small nuclear RNA transcription. A third subunit is
unrelated to any eukaryotic protein and identifies tSNAP(c) as a unique
trypanosomal transcription factor. Intriguingly, the unusual
trypanosome TATA-binding protein (TBP) tightly associates with
tSNAPc and is essential for SL RNA gene transcription. These findings
provide the first view of the architecture of a transcriptional complex
that assembles at an RNA polymerase II-dependent gene promoter in a
highly divergent eukaryote.
PMID: 16055739 [PubMed - indexed for MEDLINE]
10: Mol Cell Biol. 2005 Aug;25(16):7303-13.
Characterization of a multisubunit transcription factor complex
essential for spliced-leader RNA gene transcription in Trypanosoma
brucei.
Schimanski B, Nguyen TN, Gunzl A.
Department of Genetics and Developmental Biology, University of
Connecticut Health Center, Farmington, 06030-3710, USA.
In the unicellular human parasites Trypanosoma brucei, Trypanosoma
cruzi, and Leishmania spp., the spliced-leader (SL) RNA is a key
molecule in gene expression donating its 5'-terminal region in SL
addition trans splicing of nuclear pre-mRNA. While there is no evidence
that this process exists in mammals, it is obligatory in mRNA
maturation of trypanosomatid parasites. Hence, throughout their life
cycle, these organisms crucially depend on high levels of SL RNA
synthesis. As putative SL RNA gene transcription factors, a partially
characterized small nuclear RNA-activating protein complex (SNAP(c))
and the TATA-binding protein related factor 4 (TRF4) have been
identified thus far. Here, by tagging TRF4 with a novel epitope
combination termed PTP, we tandem affinity purified from crude T.
brucei extracts a stable and transcriptionally active complex of six
proteins. Besides TRF4 these were identified as extremely divergent
subunits of SNAP(c) and of transcription factor IIA (TFIIA). The
latter finding was unexpected since genome databases of trypanosomatid
parasites appeared to lack general class II transcription factors. As
we demonstrate, the TRF4/SNAP(c)/TFIIA complex binds specifically to
the SL RNA gene promoter upstream sequence element and is absolutely
essential for SL RNA gene transcription in vitro.
PMID: 16055739 [PubMed - indexed for MEDLINE]
11: RNA. 2005 Sep;11(9):1430-40. Epub 2005 Jul 25.
A mutational analysis of U12-dependent splice site dinucleotides.
Dietrich RC, Fuller JD, Padgett RA.
Department of Molecular Genetics, NE-20, Lerner Research Institute,
Cleveland Clinic Foundation, 9500 Euclid Ave., Cleveland, OH 44195,
USA.
Introns spliced by the U12-dependent minor spliceosome are divided into
two classes based on their splice site dinucleotides. The /AU-AC/ class
accounts for about one-third of U12-dependent introns in humans, while
the /GU-AG/ class accounts for the other two-thirds. We have
investigated the in vivo and in vitro splicing phenotypes of mutations
in these dinucleotide sequences. A 5' A residue can splice to any 3'
residue, although C is preferred. A 5' G residue can splice to 3' G or
U residues with a preference for G. Little or no splicing was observed
to 3' A or C residues. A 5' U or C residue is highly deleterious for
U12-dependent splicing, although some combinations, notably 5' U to 3'
U produced detectable spliced products. The dependence of 3' splice
site activity on the identity of the 5' residue provides evidence for
communication between the first and last nucleotides of the intron.
Most mutants in the second position of the 5' splice site and the next
to last position of the 3' splice site were defective for splicing.
Double mutants of these residues showed no evidence of communication
between these nucleotides. Varying the distance between the branch site
and the 3' splice site dinucleotide in the /GU-AG/ class showed that a
somewhat larger range of distances was functional than for the /AU-AC/
class. The optimum branch site to 3' splice site distance of 11-12
nucleotides appears to be the same for both classes.
PMID: 16043500 [PubMed - indexed for MEDLINE]
12: IUBMB Life. 2005 Mar;57(3):173-9.
A novel snoRNA can direct site-specific 2'-O-ribose methylation of
snRNAs in Oryza sativa.
Li W, Jiang G, Huang B, Jin Y.
Shanghai Institutes for Biological Sciences, Chinese Academy of
Sciences, Shanghai, China.
Small nucleolar RNAs (snoRNAs) are a kind of noncoding RNAs, and the
vast majority of snoRNAs are involved in site-specific modifications of
rRNAs. A novel box C/D snoRNA called snoR124 was found inOryza sativa,
and it can direct 2'-O-ribose methylation of spliceosomal small nuclear
RNAs (snRNAs). The snoRNA has two antisense elements, and the results
of primer extensions at different dNTP concentrations provide evidence
that snoR124 guide 2'-O-methylations of the C76 residue in the U4 snRNA
and the T91 residue in the U5 snRNA. In addition, this snoRNA is
located in a snoRNA gene cluster with another 7 snoRNAs which are
identified to direct ribose methylations in rRNAs. This is consistent
with the opinion that the snoRNA gene organization in plant is mainly
gene cluster. The snoR124 is the first example of a snoRNA that directs
modifications of RNAs other than rRNAs in plant; it will avail to get
more insights into the function of snoRNAs in plant.
PMID: 16036579 [PubMed - indexed for MEDLINE]
13: Cancer Res. 2005 Jul 15;65(14):6228-36.
CaSm-mediated cellular transformation is associated with altered gene
expression and messenger RNA stability.
Fraser MM, Watson PM, Fraig MM, Kelley JR, Nelson PS, Boylan AM, Cole
DJ, Watson DK.
Department of Pathology and Laboratory Medicine, Hollings Cancer
Center, Medical University of South Carolina, Charleston, South
Carolina 29403, USA.
CaSm (cancer-associated Sm-like) was originally identified based on
elevated expression in pancreatic cancer and in several cancer-derived
cell lines. CaSm encodes a 133 amino acid protein that contains two Sm
motifs found in the common small nuclear RNA proteins and the LSm
(like-Sm) family of proteins. Compared with normal human prostate
tissue and primary prostate epithelial cells, some primary prostate
tumors and prostate cancer-derived cell lines have elevated CaSm
expression. Expression of antisense CaSm RNA in DU145 cells results in
reduced CaSm protein levels and less transformed phenotype, measured by
anchorage-independent growth in vitro and tumor formation in severe
combined immunodeficient mice in vivo. Additional data shows that
adenoviral delivery of antisense CaSm inhibits the growth of prostate
cancer cell lines by altering cell cycle progression, and is associated
with reduced expression of cyclin B1 and CDK1 proteins. Consistent with
failure of antisense-treated cells to enter mitosis, microarray
analysis identified altered expression of NEK2 and nucleophosmin/B23.
Although the mechanisms by which CaSm contributes to neoplastic
transformation and cellular proliferation are unknown, it has been
shown that the yeast homologue (spb8/LSm1) of CaSm is required for 5'
to 3' degradation of specific mRNAs. We provide data consistent with a
similar role for CaSm in human cells, supporting the hypothesis that
elevated CaSm expression observed in cancer leads to destabilization of
multiple gene transcripts, contributing to the mutator phenotype of
cancer cells.
PMID: 16036579 [PubMed - indexed for MEDLINE]
14: J Cutan Pathol. 2005 Aug;32(7):474-83.
Epstein--Barr virus-associated B-cell lymphoma in the setting of
iatrogenic immune dysregulation presenting initially in the skin.
Verma S, Frambach GE, Seilstad KH, Nuovo G, Porcu P, Magro CM.
School of Medicine and Public Health, The Ohio State University,
Columbus, OH, USA.
BACKGROUND: Epstein-Barr virus (EBV) has been implicated in B-cell
lymphoma associated with iatrogenic immune dysregulation, primarily in
the context of extracutaneous lymphoma. METHODS: We describe six
patients, five transplant recipients receiving cyclosporine and one
patient with rheumatoid arthritis receiving methotrexate, who developed
cutaneous presentations of EBV-associated B-cell lymphoma. Human
herpesvirus 8 (HHV8) and EBV thymidine kinase (vTK) expression were
also explored. RESULTS: The cases comprised plasmablastic lymphoma (one
case), plasmacytic marginal zone lymphoma (two cases), and diffuse
large B-cell lymphoma (three cases). There was a monoclonal gammopathy
in one and concurrent extracutaneous disease in two of the six
patients. EBV-associated latent small nuclear RNA was detected in all
cases with coexpression of HHV8 in one of the five cases and of vTK in
three of the six cases. Three patients responded to a reduction in the
immunosuppressive regimen and antiviral therapy. Recurrent disease
developed in two, with one patient succumbing to multiorgan
dissemination. CONCLUSIONS: EBV-associated cutaneous B-cell lymphoma is
characterized by a long interval between the initiation of
immunosuppression and the development of lymphoma. Although previous
reports have reported an indolent clinical course, an aggressive
clinical course may occur. HHV8 and lytic phase EBV antigens are
detected in some cases, possibly suggesting a pathogenetic role.
PMID: 16008691 [PubMed - indexed for MEDLINE]
15: J Biol Chem. 2005 Aug 26;280(34):30619-29. Epub 2005 Jun 30.
Transcription-dependent association of multiple positive transcription
elongation factor units to a HEXIM multimer.
Dulac C, Michels AA, Fraldi A, Bonnet F, Nguyen VT, Napolitano G, Lania
L, Bensaude O.
Unite Mixte de Recherche 8541 CNRS, Ecole Normale Superieure,
Laboratoire de Regulation de l'Expression Genetique, 75230 Paris Cedex
05, France.
The positive transcription elongation factor (P-TEFb) comprises a
kinase, CDK9, and a Cyclin T1 or T2. Its activity is inhibited by
association with the HEXIM1 or HEXIM2 protein bound to 7SK small
nuclear RNA. HEXIM1 and HEXIM2 were found to form stable homo- and
hetero-oligomers. Using yeast two-hybrid and transfection assays, we
have now shown that the C-terminal domains of HEXIM proteins directly
interact with each other. Hydrodynamic parameters measured by glycerol
gradient ultracentrifugation and gel-permeation chromatography
demonstrate that both purified recombinant and cellular HEXIM1 proteins
form highly anisotropic particles. Chemical cross-links suggest that
HEXIM1 proteins form dimers. The multimeric nature of HEXIM1 is
maintained in P-TEFb.HEXIM1.7SK RNA complexes. Multiple P-TEFb modules
are found in the inactive P-TEFb.HEXIM1.7SK complexes. It is proposed
that 7SK RNA binding to a HEXIM1 multimer promotes the simultaneous
recruitment and hence inactivation of multiple P-TEFb units.
PMID: 15994294 [PubMed - indexed for MEDLINE]
*****THROWN OUT –NO ABSTRACT*****
16: Methods Mol Biol. 2005;309:321-32.
Modification of human U1 snRNA for inhibition of gene expression at the
level of pre-mRNA.
Liu P, Stover ML, Lichtler A, Rowe DW.
Department of Genetics and Developmental Biology, University of
Connecticut Health Center, Farmington, USA.
PMID: 15990411 [PubMed - indexed for MEDLINE]
*****END THROW OUT*****
17: Mol Cell. 2005 Jul 1;19(1):65-75.
Cotranscriptional spliceosome assembly dynamics and the role of U1
snRNA:5'ss base pairing in yeast.
Lacadie SA, Rosbash M.
Howard Hughes Medical Institute, Biology Department MS008, Brandeis
University, 415 South Street, Waltham, Massachusetts 02454, USA.
To investigate the mechanism of spliceosome assembly in vivo, we
performed chromatin immunoprecipitation (ChIP) analysis of U1, U2, and
U5 small nuclear ribonucleoprotein particles (snRNPs) to introncontaining yeast (S. cerevisiae) genes. The snRNPs display patterns
that indicate a cotranscriptional assembly model: U1 first, then U2,
and the U4/U6*U5 tri-snRNP followed by U1 destabilization. cis-splicing
mutations also support a role of U2 and/or the tri-snRNP in U1
destabilization. Moreover, they indicate that splicing efficiency has a
major impact on cotranscriptional snRNP recruitment and suggest that
cotranscriptional recruitment of U2 or the tri-snRNP is required to
commit the pre-mRNA to splicing. Branchpoint (BP) mutations had a major
effect on the U1 pattern, whereas 5' splice site (5'ss) mutations had a
stronger effect on the U2 pattern. A 5'ss-U1 snRNA complementation
experiment suggests that pairing between U1 and the 5'ss occurs after
U1 recruitment and contributes to a specific U1:substrate conformation
required for efficient U2 and tri-snRNP recruitment.
PMID: 15989965 [PubMed - indexed for MEDLINE]
18: Mol Cell Biol. 2005 Jul;25(13):5543-51.
The survival of motor neurons protein determines the capacity for snRNP
assembly: biochemical deficiency in spinal muscular atrophy.
Wan L, Battle DJ, Yong J, Gubitz AK, Kolb SJ, Wang J, Dreyfuss G.
Howard Hughes Medical Institute, Department of Biochemistry &
Biophysics, University of Pennsylvania School of Medicine,
Philadelphia, Pennsylvania 19104-6148, USA.
Reduction of the survival of motor neurons (SMN) protein levels causes
the motor neuron degenerative disease spinal muscular atrophy, the
severity of which correlates with the extent of reduction in SMN. SMN,
together with Gemins 2 to 7, forms a complex that functions in the
assembly of small nuclear ribonucleoprotein particles (snRNPs).
Complete depletion of the SMN complex from cell extracts abolishes
snRNP assembly, the formation of heptameric Sm cores on snRNAs.
However, what effect, if any, reduction of SMN protein levels, as
occurs in spinal muscular atrophy patients, has on the capacity of
cells to produce snRNPs is not known. To address this, we developed a
sensitive and quantitative assay for snRNP assembly, the formation of
high-salt- and heparin-resistant stable Sm cores, that is strictly
dependent on the SMN complex. We show that the extent of Sm core
assembly is directly proportional to the amount of SMN protein
in cell extracts. Consistent with this, pulse-labeling experiments
demonstrate a significant reduction in the rate of snRNP biogenesis in
low-SMN cells. Furthermore, extracts of cells from spinal muscular
atrophy patients have a lower capacity for snRNP assembly that
corresponds directly to the reduced amount of SMN. Thus, SMN determines
the capacity for snRNP biogenesis, and our findings provide evidence
for a measurable deficiency in a biochemical activity in cells from
patients with spinal muscular atrophy.
PMID: 15964810 [PubMed - indexed for MEDLINE]
19: Mol Cell Biol. 2005 Jul;25(13):5523-34.
Esf2p, a U3-associated factor required for small-subunit processome
assembly and compaction.
Hoang T, Peng WT, Vanrobays E, Krogan N, Hiley S, Beyer AL, Osheim YN,
Greenblatt J, Hughes TR, Lafontaine DL.
Fonds National de la Recherche Scientifique, Universite Libre de
Bruxelles, Institut de Biologie et de Medecine Moleculaires, CharleroiGosselies, Belgium.
Esf2p is the Saccharomyces cerevisiae homolog of mouse ABT1, a protein
previously identified as a putative partner of the TATA-element binding
protein. However, large-scale studies have indicated that Esf2p is
primarily localized to the nucleolus and that it physically associates
with pre-rRNA processing factors. Here, we show that Esf2p-depleted
cells are defective for pre-rRNA processing at the early nucleolar
cleavage sites A0 through A2 and consequently are inhibited for 18S
rRNA synthesis. Esf2p was stably associated with the 5' external
transcribed spacer (ETS) and the box C+D snoRNA U3, as well as
additional box C+D snoRNAs and proteins enriched within the smallsubunit (SSU) processome/90S preribosomes. Esf2p colocalized on
glycerol gradients with 90S preribosomes and slower migrating particles
containing 5' ETS fragments. Strikingly, upon Esf2p depletion,
chromatin spreads revealed that SSU processome assembly and compaction
are inhibited and glycerol gradient analysis showed that U3 remains
associated within 90S preribosomes. This suggests that in the absence
of proper SSU processome assembly, early pre-rRNA processing is
inhibited and U3 is not properly released from the 35S pre-rRNAs. The
identification of ABT1 in a large-scale analysis of the human nucleolar
proteome indicates that its role may also be conserved in mammals.
PMID: 15964808 [PubMed - indexed for MEDLINE]
20: EMBO J. 2005 Jul 6;24(13):2403-13. Epub 2005 Jun 16.
Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNAguided or RNA-independent mechanism.
Ma X, Yang C, Alexandrov A, Grayhack EJ, Behm-Ansmant I, Yu YT.
Department of Biochemistry and Biophysics, University of Rochester
Medical Center, Rochester, NY 14642, USA.
Yeast U2 small nuclear RNA (snRNA) contains three pseudouridines
(Psi35, Psi42, and Psi44). Pus7p and Pus1p catalyze the formation of
Psi35 and Psi44, respectively, but the mechanism of Psi42 formation
remains unclear. Using a U2 substrate containing a single (32)P
radiolabel at position 42, we screened a GST-ORF library for
pseudouridylase activity. Surprisingly, we found a Psi42-specific
pseudouridylase activity that coincided with Nhp2p, a protein
component of a Box H/ACA sno/scaRNP (small nucleolar/Cajal bodyspecific ribonucleoprotein). When isolated by tandem affinity
purification (TAP), the other protein components of the H/ACA
sno/scaRNP also copurified with the pseudouridylase activity.
Micrococcal nuclease-treated TAP preparations were devoid of
pseudouridylase activity; however, activity was restored upon addition
of RNAs from TAP preparations. Pseudouridylation reconstitution using
RNAs from a Box H/ACA RNA library identified snR81, a snoRNA known to
guide rRNA pseudouridylation, as the Psi42-specific guide RNA. Using
the snR81-deletion strain, Nhp2p- or Cbf5p-conditional depletion
strain, and a cbf5 mutation strain, we further demonstrated that the
pseudouridylase activity is dependent on snR81 snoRNP in vivo. Our data
indicate that snRNA pseudouridylation can be catalyzed by both RNAdependent and RNA-independent mechanisms.
PMID: 15962000 [PubMed - indexed for MEDLINE]
21: Nucleic Acids Res. 2005 Jun 13;33(10):3435-46. Print 2005.
Loss of G-A base pairs is insufficient for achieving a large opening of
U4 snRNA K-turn motif.
Cojocaru V, Klement R, Jovin TM.
Department of Molecular Biology, Max Planck Institute for Biophysical
Chemistry Am Fassberg 11, 37077 Gottingen, Germany.
Upon binding to the 15.5K protein, two tandem-sheared G-A base pairs
are formed in the internal loop of the kink-turn motif of U4 snRNA (KtU4). We have reported that the folding of Kt-U4 is assisted by protein
binding. Unstable interactions that contribute to a large opening of
the free RNA ('k-e motion') were identified using locally enhanced
sampling molecular dynamics simulations, results that agree with
experiments. A detailed analysis of the simulations reveals that the ke motion in Kt-U4 is triggered both by loss of G-A base pairs in the
internal loop and backbone flexibility in the stems. Essential dynamics
show that the loss of G-A base pairs is correlated along the first mode
but anti-correlated along the third mode with the k-e motion. Moreover,
when enhanced sampling was confined to the internal loop, the RNA
adopted an alternative conformation characterized by a sharper kink,
opening of G-A base pairs and modified stacking interactions. Thus,
loss of G-A base pairs is insufficient for achieving a large opening of
the free RNA. These findings, supported by previously published RNA
structure probing experiments, suggest that G-A base pair formation
occurs upon protein binding, thereby stabilizing a selective
orientation of the stems.
PMID: 15956103 [PubMed - indexed for MEDLINE]
22: J Biol Chem. 2005 Jul 29;280(30):27697-704. Epub 2005 Jun 14.
Cooperation between small nuclear RNA-activating protein complex
(SNAPC) and TATA-box-binding protein antagonizes protein kinase CK2
inhibition of DNA binding by SNAPC.
Gu L, Esselman WJ, Henry RW.
Department of Biochemistry and Molecular Biology, Michigan State
University, East Lansing, Michigan 48824, USA.
Protein kinase CK2 regulates RNA polymerase III transcription of human
U6 small nuclear RNA (snRNA) genes both negatively and positively
depending upon whether the general transcription machinery or RNA
polymerase III is preferentially phosphorylated. Human U1 snRNA genes
share similar promoter architectures as that of U6 genes but are
transcribed by RNA polymerase II. Herein, we report that CK2 inhibits
U1 snRNA gene transcription by RNA polymerase II. Decreased levels of
endogenous CK2 correlates with increased U1 expression, whereas CK2
associates with U1 gene promoters, indicating that it plays a direct
role in U1 gene regulation. CK2 phosphorylates the general
transcription factor small nuclear RNA-activating protein complex
(SNAP(C)) that is required for both RNA polymerase II and III
transcription, and SNAP(C) phosphorylation inhibits binding to snRNA
gene promoters. However, restricted promoter access by phosphorylated
SNAP(C) can be overcome by cooperative interactions with TATA-boxbinding protein at a U6 promoter but not at a U1 promoter. Thus, CK2
may have the capacity to differentially regulate U1 and U6
ranscription even though SNAP(C) is universally utilized for human
snRNA gene transcription.
PMID: 15955816 [PubMed - indexed for MEDLINE]
23: Eukaryot Cell. 2005 Jun;4(6):1155-7.
Small sense and antisense RNAs derived from a telomeric retroposon
family in Giardia intestinalis.
Ullu E, Lujan HD, Tschudi C.
Department of Internal Medicine, Yale Medical School, BCMM 136D, 295
Congress Avenue, Box 9812, New Haven, CT 06536-8012, USA.
elisabetta.ullu@yale.edu
Sequencing of a library of small RNAs from Giardia intestinalis
identified a novel class of small sense and antisense RNAs homologous
to the retroposon family GilT/Genie1 that is located at certain
telomeres. These small RNAs may contribute to silencing GilT expression
via the RNA interference pathway.
PMID: 15947207 [PubMed - indexed for MEDLINE]
24: Proc Natl Acad Sci U S A. 2005 Jun 14;102(24):8555-60. Epub 2005
Jun 7.
HEXIM1 forms a transcriptionally abortive complex with glucocorticoid
receptor without involving 7SK RNA and positive transcription
elongation factor b.
Shimizu N, Ouchida R, Yoshikawa N, Hisada T, Watanabe H, Okamoto K,
Kusuhara M, Handa H, Morimoto C, Tanaka H.
Division of Clinical Immunology and Department of Rheumatology and
Allergy, Institute of Medical Science, University of Tokyo, 4-6-1,
Shirokanedai, Minato-ku, Tokyo 108-8639, Japan.
The HEXIM1 protein has been shown to form a protein-RNA complex
composed of 7SK small nuclear RNA and positive transcription elongation
factor b (P-TEFb), which is composed of cyclin-dependent kinase 9
(CDK9) and cyclin T1, and to inhibit the kinase activity of CDK9,
thereby suppressing RNA polymerase II-dependent transcriptional
elongation. Here, we biochemically demonstrate that HEXIM1 forms
a distinct complex with glucocorticoid receptor (GR) without RNA, CDK9,
or cyclin T1. HEXIM1, through its arginine-rich nuclear localization
signal, directly associates with the ligand-binding domain of GR.
Introduction of HEXIM1 short interfering RNA and adenovirus-mediated
exogenous expression of HEXIM1 positively and negatively modulated
glucocorticoid-responsive gene activation, respectively. In the
nucleus, HEXIM1 was shown to localize in a distinct compartment from
that of the p160 coactivator transcriptional intermediary factor 2.
Overexpression of HEXIM1 decreased ligand-dependent association
between GR and transcriptional intermediary factor 2. Antisensemediated disruption of 7SK blunted the negative effect of HEXIM1 on
arylhydrocarbon receptor-dependent transcription but not on GR-mediated
one, indicating that a class of transcription factors are direct
targets of HEXIM1. These results indicate that HEXIM1 has dual roles in
transcriptional regulation: inhibition of transcriptional elongation
dependent on 7SK RNA and positive transcription elongation factor b and
interference with the sequence-specific transcription factor GR via a
direct protein-protein interaction. Moreover, the fact that the
central nuclear localization signal of HEXIM1 is essential for both of
these
actions may argue the crosstalk of these functions.
PMID: 15941832 [PubMed - indexed for MEDLINE]
25: Biochem Biophys Res Commun. 2005 Jul 22;333(1):64-9.
The effect of U1 snRNA binding free energy on the selection of 5'
splice sites.
Bi J, Xia H, Li F, Zhang X, Li Y.
MOE Key Laboratory of Bioinformatics, Department of Automation,
Tsinghua University, Beijing, China.
The importance of U1 snRNA binding free energy in the regulation of
alternative splicing has been studied in some genes with site-directed
mutagenesis. Here we report a large-scale analysis of its impact on 5'
splice site (5'ss) selection in human genome. The results show that
free energy exerts different effects on alternative 5'ss choice in
different situations and -8.1 kcal/mol is a threshold. When both free
energies of two competing 5'ss are larger than -8.1 kcal/mol, the 5'ss
with lower free energy is more frequently used. However, in other pairs
of 5'ss, lower-free-energy 5'ss does not seem to be favored and even
the other 5'ss is used more frequently, which suggests that very low
binding free energy would impair splicing. Some observations hold true
only for those alternative 5' splicing with short alternative exons
(<50nt), which implies a complex mechanism of 5'ss selection involving
both U1 snRNA binding free energy and regulatory factors.
PMID: 15936716 [PubMed - indexed for MEDLINE]
26: Mol Cell Biol. 2005 Jun;25(12):4813-25.
Proximity of the U12 snRNA with both the 5' splice site and the branch
point during early stages of spliceosome assembly.
Frilander MJ, Meng X.
Institute of Biotechnology, Program on Developmental Biology, PL56
(Viikinkaari 9), 00014 University of Helsinki, Finland.
Mikko.Frilander@Helsinki.Fi
U12 snRNA is required for branch point recognition in the U12-dependent
spliceosome. Using site-specific cross-linking, we have captured an
unexpected interaction between the 5' end of the U12 snRNA and the -2
position upstream of the 5' splice site of P120 and SCN4a splicing
substrates. The U12 snRNA nucleotides that contact the 5' exon are the
same ones that form the catalytically important helix Ib with U6atac
snRNA in the spliceosome catalytic core. However, the U12/5' exon
interaction is transient, occurring prior to the entry of the
U4atac/U6atac.U5 tri-snRNP to the spliceosome. This suggests that
the helix Ib region of U12 snRNA is positioned near the 5' splice site
early during spliceosome assembly and only later interacts with U6atac
to form helix Ib. We also provide evidence that U12 snRNA can
simultaneously interact with 5' exon sequences near 5' splice site and
the branch point sequence, suggesting that the 5' splice site and
branch point sequences are separated by <40 to 50 A in the complex A of
the U12-dependent spliceosome. Thus, no major rearrangements are
subsequently needed to position these sites for the first step of
catalysis.
PMID: 15923601 [PubMed - indexed for MEDLINE]
*****THROWN OUT – NO ABSTRACT*****
27: Mol Biochem Parasitol. 2005 Aug;142(2):248-51.
mRNA splicing in Trypanosoma brucei: branch-point mapping reveals
differences from the canonical U2 snRNA-mediated recognition.
Lucke S, Jurchott K, Hung LH, Bindereif A.
Institut fur Biochemie, Justus-Liebig-Universitat Giessen, HeinrichBuff-Ring 58, D-35392 Giessen, Germany.
PMID: 15923047 [PubMed - indexed for MEDLINE]
*****END THROW OUT*****
28: Mol Ther. 2005 Jun;11(6):899-905.
In vivo inhibition of hippocampal Ca2+/calmodulin-dependent protein
kinase II by RNA interference.
Babcock AM, Standing D, Bullshields K, Schwartz E, Paden CM, Poulsen
DJ.
Department of Psychology, Montana State University, Bozeman, MT 59717,
USA. mbabeock@montana.edu
Hippocampal alpha-Ca2+/calmodulin-dependent protein kinase II (alphaCaMKII) has been implicated in spatial learning, neuronal plasticity,
epilepsy, and cerebral ischemia. In the present study, an adenoassociated virus (AAV) vector was designed to express green fluorescent
protein (GFP) from the CBA promoter and a small hairpin RNA targeting
alpha-CaMKII (AAV-shCAM) driven from the U6 promoter. The AAV-shCAM or
control vector was microinfused into the rat hippocampus and behavioral
testing conducted 19-26 days following surgery. Expression of the
marker gene and alpha-CaMKII was evaluated 31 days following
AAV infusion. GFP expression was localized to the hippocampus and
extended +/-2 mm rostral and caudal from the injection site.
Hippocampal alpha-CaMKII was significantly reduced following AAV-shCAM
treatment as demonstrated using immunohistochemical and Western
analysis. This suppression of alpha-CaMKII was associated with changes
in exploratory behavior (open field task) and impaired place learning
(water maze task). These results demonstrate the efficacy of a viralbased delivered shRNA to produce gene suppression in a specific circuit
of the brain.
PMID: 15922960 [PubMed - indexed for MEDLINE]
29: Curr Biol. 2005 May 24;15(10):974-9.
Small nuclear RNAs encoded by Herpesvirus saimiri upregulate the
expression of genes linked to T cell activation in virally transformed
T cells.
Cook HL, Lytle JR, Mischo HE, Li MJ, Rossi JJ, Silva DP, Desrosiers RC,
Steitz JA.
Howard Hughes Medical Institute, Department of Molecular Biophysics and
Biochemistry, Yale University, New Haven, Connecticut 06536, USA.
Seven small nuclear RNAs of the Sm class are encoded by Herpesvirus
saimiri (HVS), a gamma Herpesvirus that causes aggressive T cell
leukemias and lymphomas in New World primates and efficiently
transforms T cells in vitro. The Herpesvirus saimiri U RNAs (HSURs) are
the most abundant viral transcripts in HVS-transformed, latently
infected T cells but are not required for viral replication or
transformation in vitro. We have compared marmoset T cells
transformed with wild-type or a mutant HVS lacking the most highly
conserved HSURs, HSURs 1 and 2. Microarray and Northern analyses reveal
that HSUR 1 and 2 expression correlates with significant increases in a
small number of host mRNAs, including the T cell-receptor beta and
gamma chains, the T cell and natural killer (NK) cell-surface receptors
CD52 and DAP10, and intracellular proteins--SKAP55, granulysin, and
NKG7--linked to T cell and NK cell activation. Upregulation of three of
these transcripts was rescued after transduction of deletion-mutantHVS-transformed cells with a lentiviral vector carrying HSURs 1
and 2. These changes indicate an unexpected role for the HSURs in
regulating a remarkably defined and physiologically relevant set of
host targets involved in the activation of virally transformed T cells
during latency.
PMID: 15916956 [PubMed - indexed for MEDLINE]
30: Biochem Soc Trans. 2005 Jun;33(Pt 3):447-9.
Towards understanding the catalytic core structure of the spliceosome.
Butcher SE, Brow DA.
Departments of Biochemistry and Biomolecular Chemistry, University of
Wisconsin-Madison, Madison, WI 53706, USA. butcher@nmrfam.wisc.edu
The spliceosome catalyses the splicing of nuclear pre-mRNA (precursor
mRNA) in eukaryotes. Pre-mRNA splicing is essential to remove internal
non-coding regions of pre-mRNA (introns) and to join the remaining
segments (exons) into mRNA before translation. The spliceosome is a
complex assembly of five RNAs (U1, U2, U4, U5 and U6) and many dozens
of associated proteins. Although a high-resolution structure of the
spliceosome is not yet available, inroads have been made towards
understanding its structure and function. There is growing evidence
suggesting that U2 and U6 RNAs, of the five, may contribute to the
catalysis of pre-mRNA splicing. In this review, recent progress towards
understanding the structure and function of U2 and U6 RNAs is
summarized. Publication Types:
Review
Review, Tutorial
PMID: 15916538 [PubMed - indexed for MEDLINE]
31: Hum Gene Ther. 2005 May;16(5):618-26.
Adenovirus-mediated silencing of huntingtin expression by shRNA.
Huang B, Kochanek S.
Division of Gene Therapy, University of Ulm, D-89081 Ulm, Germany.
Huntington's disease (HD) is an inherited autosomal dominant,
neurodegenerative disease that is caused by a gain of function mutation
characterized by the expansion of a CAG trinucleotide repeat in exon 1
of the huntingtin (htt) gene. Since hairpin small interference RNA
(shRNA) technology allows inhibition of specific gene expression in
vitro and in vivo, vector-mediated expression of an shRNA directed to
htt mRNA could form the basis of a new treatment modality for HD. By
initial plasmid transfection of 293 cells, we identified one exon
1-targeted shRNA, which efficiently inhibited expression of an htt exon
1-GFP fusion protein and the endogenous htt gene. A replicationdeficient adenovirus (Ad) vector Adie-1-1 was constructed to express
this shRNA from the U6 promoter. In A549 cells expressing exon 1 of htt
with an expanded CAG allele, Adie- 1-1 efficiently prevented htt exon 1
expression and htt aggregate formation. In addition, in different
neuronal and nonneuronal cell lines, Adie-1-1 efficiently
inhibited the expression of endogenous htt. Together, this data
indicates the delineation of an shRNA strategy that may become the
basis for treatment of HD.
PMID: 15916486 [PubMed - indexed for MEDLINE]
32: Nucleic Acids Res. 2005 May 24;33(9):2917-28. Print 2005.
Kinetic analysis of the role of the tyrosine 13, phenylalanine 56 and
glutamine 54 network in the U1A/U1 hairpin II interaction.
Law MJ, Chambers EJ, Katsamba PS, Haworth IS, Laird-Offringa IA.
Department of Biochemistry and Molecular Biology, University of
Southern California Los Angeles, CA 90089-9176, USA.
The A protein of the U1 small nuclear ribonucleoprotein particle,
interacting with its stem-loop RNA target (U1hpII), is frequently used
as a paradigm for RNA binding by recognition motif domains (RRMs).
U1A/U1hpII complex formation has been proposed to consist of at least
two steps: electrostatically mediated alignment of both molecules
followed by locking into place, based on the establishment of closerange interactions. The sequence of events between alignment and
locking remains obscure. Here we examine the roles of three critical
residues, Tyr13, Phe56 and Gln54, in complex formation and stability
using Biacore. Our mutational and kinetic data suggest that Tyr13 plays
a more important role than Phe56 in complex formation. Mutational
analysis of Gln54, combined with molecular dynamics studies, points to
Arg52 as another key residue in association. Based on our data and
previous structural and modeling studies, we propose that electrostatic
alignment of the molecules is followed by hydrogen bond formation
between the RNA and Arg52, and the sequential establishment of
interactions with loop bases (including Tyr13). A quadruple stack,
sandwiching two bases between Phe56 and Asp92, would occur last and
coincide with the rearrangement of a C-terminal helix that partially
occludes the RRM surface in the free protein.
PMID: 15914668 [PubMed - indexed for MEDLINE]
33: Cell. 2005 May 20;121(4):529-39.
Comment in: Cell. 2005 May 20;121(4):495-6.
Structural insights into RNA quality control: the Ro autoantigen binds
misfolded RNAs via its central cavity.
Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM.
Department of Cell Biology, Yale University School of Medicine, New
Haven, CT 06510, USA.
The Ro 60 kDa autoantigen is a major target of the immune response in
patients with systemic lupus erythematosus. In vertebrate cells, Ro
binds misfolded small RNAs and likely functions in RNA quality control.
In eukaryotes and bacteria, Ro also associates with small RNAs called Y
RNAs. We present structures of unliganded Ro and Ro complexed with two
RNAs at 1.95 and 2.2 A resolution, respectively. Ro consists of a von
Willebrand factor A domain and a doughnut-shaped domain composed of
HEAT repeats. In the complex, a fragment of Y RNA binds on the outer
surface of the HEAT-repeat ring, and single-stranded RNA binds in the
toroid hole. Mutagenesis supports a binding site for misfolded RNAs
that encompasses both sites, with a single-stranded end inserted into
the toroid cavity. Our experiments suggest that one role of Y RNAs may
be to regulate access of other RNAs to Ro.
PMID: 15907467 [PubMed - indexed for MEDLINE]
34: Cell. 2005 May 20;121(4):495-6.
Comment on: Cell. 2005 May 20;121(4):529-39.
Ro's role in RNA reconnaissance.
Macrae IJ, Doudna JA.
Howard Hughes Medical Institute, Department of Molecular & Cell
Biology, Lawrence Berkeley National Laboratory, University of
California at Berkeley, Berkeley, CA 94720, USA.
The Ro 60 kDa autoantigen binds misfolded RNAs and likely functions in
small RNA quality control. In this issue of Cell, Stein et al. (2005)
present crystal structures of Ro alone and bound to both double- and
single-stranded RNA, revealing two distinct RNA binding sites that
suggest how Ro may distinguish between native and misfolded small RNAs.
Publication Types: Comment
Review
Review, Tutorial
PMID: 15907458 [PubMed - indexed for MEDLINE]
35: J Biol Chem. 2005 Jul 8;280(27):25478-84. Epub 2005 May 16.
Substrate-dependent differences in U2AF requirement for splicing in
adenovirus-infected cell extracts.
Lutzelberger M, Backstrom E, Akusjarvi G.
Department of Medical Biochemistry and Microbiology, Uppsala
University, 75123Uppsala, Sweden.
U2AF has been characterized as an essential splicing factor required
for efficient recruitment of U2 small nuclear ribonucleoprotein to the
3'-splice site in a pre-mRNA. The U2AF65 subunit binds to the
pyrimidine tract of the pre-mRNA, whereas the U2AF(35) subunit contacts
the 3'-splice site AG. Here we show that U2AF35 appears to be
completely dispensable for splicing in nuclear extracts prepared from
adenovirus late-infected cells (Ad-NE). As a consequence, the viral
IIIa and cellular IgM introns, which both have suboptimal 3'-splice
sites and require U2AF35 for splicing in nuclear extracts from
uninfected cells, are transformed to U2AF35-independent introns in AdNE. Furthermore, we present evidence that two parallel pathways of 3'splice site recognition exist in Ad-NE. We show that the viral 52,55K
intron, which has an extended pyrimidine tract, requires U2AF for
activity in Ad-NE. In contrast, the IgM intron, which has a weak 3'splice site sequence context, undergoes the first catalytic step
of splicing in U2AF-depleted Ad-NE, suggesting that spliceosome
assembly occurs through a novel U2AF-independent pathway in Ad-NE.
PMID: 15899895 [PubMed - indexed for MEDLINE]
36: Nucleic Acids Res. 2005 May 13;33(9):2781-91. Print 2005.
Unusual features of fibrillarin cDNA and gene structure in Euglena
gracilis: evolutionary conservation of core proteins and structural
predictions for methylation-guide box C/D snoRNPs throughout the domain
Eucarya.
Russell AG, Watanabe Y, Charette JM, Gray MW.
Department of Biochemistry and Molecular Biology, Dalhousie University
Halifax, Nova Scotia, Canada B3H 1X5. russella@dal.ca
Box C/D ribonucleoprotein (RNP) particles mediate O2'-methylation of
rRNA and other cellular RNA species. In higher eukaryotic taxa, these
RNPs are more complex than their archaeal counterparts, containing four
core protein components (Snu13p, Nop56p, Nop58p and fibrillarin)
compared with three in Archaea. This increase in complexity raises
questions about the evolutionary emergence of the eukaryote-specific
proteins and structural conservation in these RNPs throughout the
eukaryotic domain. In protists, the primarily unicellular organisms
comprising the bulk of eukaryotic diversity, the protein composition of
box C/D RNPs has not yet been extensively explored. This study
describes the complete gene, cDNA and protein sequences of the
fibrillarin homolog from the protozoon Euglena gracilis, the first such
information to be obtained for a nucleolus-localized protein in this
organism. The E.gracilis fibrillarin gene contains a mixture of intron
types exhibiting markedly different sizes. In contrast to most other
E.gracilis mRNAs characterized to date, the fibrillarin mRNA lacks a
spliced leader (SL) sequence. The predicted fibrillarin protein
sequence itself is unusual in that it contains a glycine-lysine (GK)rich domain at its N-terminus rather than the glycine-arginine-rich
(GAR) domain found in most other eukaryotic fibrillarins. In an
evolutionarily diverse collection of protists that includes E.gracilis,
we have also identified putative homologs of the other core protein
components of box C/D RNPs, thereby providing evidence that the protein
composition seen in the higher eukaryotic complexes was established
very early in eukaryotic cell evolution.
PMID: 15894796 [PubMed - indexed for MEDLINE]
41: Nucleic Acids Res. 2005 Apr 29;33(8):2493-503. Print 2005.
U1 small nuclear RNP from Trypanosoma brucei: a minimal U1 snRNA with
unusual protein components.
Palfi Z, Schimanski B, Gunzl A, Lucke S, Bindereif A.
Institut fur Biochemie, Justus-Liebig-Universitat Giessen HeinrichBuff-Ring 58, D-35392 Giessen, Germany.
Processing of primary transcripts in trypanosomes requires trans
splicing and polyadenylation, and at least for the poly(A) polymerase
gene, also internal cis splicing. The trypanosome U1 snRNA, which is
most likely a cis-splicing specific component, is unusually short and
has a relatively simple secondary structure. Here, we report the
identification of three specific protein components of the Trypanosoma
brucei U1 snRNP, based on mass spectrometry and confirmed by in vivo
epitope tagging and in vitro RNA binding. Both T.brucei U1-70K and U1C
are only distantly related to known counterparts from other eukaryotes.
The T.brucei U1-70K protein represents a minimal version of 70K,
recognizing the first loop sequence of U1 snRNA with the same
specificity as the mammalian protein. The trypanosome U1C-like protein
interacts with 70K directly and binds the 5' terminal sequence of U1
snRNA. Surprisingly, instead of U1A we have identified a novel U1
snRNP-specific protein, TbU1-24K. U1-24K lacks a known RNA-binding
motif and integrates in the U1 snRNP via interaction with U1-70K. These
data result in a model of the trypanosome U1 snRNP, which deviates
substantially from our classical view of the U1 particle and may
reflect the special requirements for splicing of a small set of cisintrons in trypanosomes.
PMID: 15863726 [PubMed - indexed for MEDLINE]
Keyword Search: “Ribosomal RNA”
1: Mol Genet Genomics. 2005 Oct 18;:1-11 [Epub ahead of print]
DEG1, encoding the tRNA:pseudouridine synthase Pus3p, impacts HOT1stimulated recombination in Saccharomyces cerevisiae.
Hepfer CE, Arnold-Croop S, Fogell H, Steudel KG, Moon M, Roff A,
Zaikoski S, Rickman A, Komsisky K, Harbaugh DL, Lang GI, Keil RL.
Department of Biology, Millersville University, 50 East Frederick
Street, PO Box 1002, Millersville, PA, 17551, USA,
carol.hepfer@millersville.edu.
In Saccharomyces cerevisiae, HOT1-stimulated recombination has been
implicated in maintaining homology between repeated ribosomal RNA
genes. The ability of HOT1 to stimulate genetic exchange requires RNA
polymerase I transcription across the recombining sequences. The transacting nuclear mutation hrm3-1 specifically reduces HOT1-dependent
recombination and prevents cell growth at 37 degrees . The HRM3 gene is
identical to DEG1. Excisive, but not gene replacement, recombination is
reduced in HOT1-adjacent sequences in deg1Delta mutants. Excisive
recombination within the genomic rDNA repeats is also decreased. The
hypo-recombination and temperature-sensitive phenotypes of deg1Delta
mutants are recessive. Deletion of DEG1 did not affect the rate of
transcription from HOT1 or rDNA suggesting that while transcription is
necessary it is not sufficient for HOT1 activity. Pseudouridine
synthase 3 (Pus3p), the DEG1 gene product, modifies the anticodon arm
of transfer RNA at positions 38 and 39 by catalyzing the conversion of
uridine to pseudouridine. Cells deficient in pseudouridine synthases
encoded by PUS1, PUS2 or PUS4 displayed no recombination defects,
indicating that Pus3p plays a specific role in HOT1 activity. Pus3p is
unique in its ability to modulate frameshifting and readthrough events
during translation, and this aspect of its activity may be
responsible for HOT1 recombination phenotypes observed in deg1 mutants.
PMID: 16231152 [PubMed - as supplied by publisher]
2: J Infect. 2005 Oct;51(3):e159-61. Epub 2005 Jan 28.
Peritonitis caused by Aspergillus sydowii in a patient undergoing
continuous ambulatory peritoneal dialysis.
Chiu YL, Liaw SJ, Wu VC, Hsueh PR.
Department of Internal Medicine, National Taiwan University Hospital,
National Taiwan University College of Medicine, No. 7, Chung-Shan South
Road, Taipei 100, Taiwan, ROC.
We describe a patient who received continuous ambulatory peritoneal
dialysis and developed catheter-related peritonitis caused by
Aspergillus sydowii. Identification of A. sydowii was confirmed by the
sequence analysis of the ribosomal RNA genes. The patient remained well
after removal of the peritoneal catheter without administration of any
anti-fungal agent.
PMID: 16230197 [PubMed - in process]
3: Br J Haematol. 2005 Nov;131(3):403-9.
Amplified-fragment length polymorphism analysis of Propionibacterium
isolates implicated in contamination of blood products.
Mohammadi T, Reesink HW, Pietersz RN, Vandenbroucke-Grauls CM,
Savelkoul PH.
Sanquin Blood Bank North West Region, Amsterdam, the Netherlands.
Propionibacterium acnes is implicated in most cases of bacterial
contamination of platelet concentrates (PCs). To determine the source
of contamination, amplified-fragment length polymorphism (AFLP)
analysis was applied. This DNA fingerprinting technique was used to
study the molecular relationship of 44 isolates derived from 22 PCs and
22 corresponding red blood cells concentrates (RBCs) from the same
whole blood donations. The AFLP results together with sequencing
analysis of the 1200 bp of the 16S ribosomal RNA gene revealed the
existence of three main groups: two groups (groups 2 and 3) (55%)
consisted of isolates that did not originate from skin flora and
another group (group 1) (45%) comprised bacteria belonging to the skin
flora. This latter group showed complete homology with reference
strains of P. acnes. Therefore these isolates can be considered as P.
acnes strains. In contrast, contaminants from groups 2 and 3 were shown
to be molecularly unrelated to the P. acnes found on the skin surface.
The AFLP is reproducible and gave invaluable information about the
nature of Propionibacteria contaminating PCs. To gain more insights
into the source of contamination, this technique could be exploited in
further studies to determine the molecular relationship of different
bacteria commonly found in blood products.
PMID: 16225662 [PubMed - in process]
4: Philos Trans R Soc Lond B Biol Sci. 2005 Oct 27;360(1462):1917-24.
Reverse taxonomy: an approach towards determining the diversity of
meiobenthic organisms based on ribosomal RNA signature sequences.
Markmann M, Tautz D.
University of Munich Zoological Institute Luissentrasse 14, 80333
Munich, Germany.
Organisms living in or on the sediment layer of water bodies constitute
the benthos fauna, which is known to harbour a large number of species
of diverse taxonomic groups. The benthos plays a significant role in
the nutrient cycle and it is, therefore, of high ecological relevance.
Here, we have explored a DNA-taxonomic approach to access the
meiobenthic organismic diversity, by focusing on obtaining signature
sequences from a part of the large ribosomal subunit rRNA (28S), the
D3-D5 region. To obtain a broad representation of taxa, benthos samples
were taken from 12 lakes in Germany, representing different ecological
conditions. In a first approach, we have extracted whole DNA from these
samples, amplified the respective fragment by PCR, cloned the fragments
and sequenced individual clones. However, we found a relatively large
number of recombinant clones that must be considered PCR artefacts. In
a second approach we have, therefore, directly sequenced PCR fragments
that were obtained from DNA extracts of randomly picked individual
organisms. In total, we have obtained 264 new unique sequences, which
can be readily placed into taxon groups, based on phylogenetic
comparison with currently available database sequences. The group
with the highest taxon abundance were nematodes and protozoa, followed
by chironomids. However, we find also that we have by far not exhausted
the diversity of organisms in the samples. Still, our data provide a
framework within which a meiobenthos DNA signature sequence database
can be constructed, that will allow to develop the necessary techniques
for studying taxon diversity in the context of ecological analysis.
Since many taxa in our analysis are initially only identified via their
signature sequences, but not yet their morphology, we propose to call
this approach 'reverse taxonomy'.
PMID: 16214749 [PubMed - in process]
5: Huan Jing Ke Xue. 2005 Jul;26(4):171-6.
[Microbial community structure analyzed by single-strand conformation
polymorphism technique in sulfate-reducing reactor]
[Article in Chinese]
Zhao YG, Ren NQ, Wang AJ, Liu GM, Zhao QS, Shang HX.
School of Municipal and Environmental Engineering, Harbin Institute of
Technology, Harbin 150090, China. zhaoyangguo@hit.edu.cn
Analyses of microbial community structure and the relationships between
Sulfate-Reducing Bacteria (SRBs) and Acidogenic Bacteria (ABs) in a
completely stirred sulfate-reducing reactor were carried out by
modified polymerase chain reaction-single-stranded conformation
polymorphism (PCR-SSCP) targeted eubacterial 16S ribosomal RNA gene. A
total of 13 bands were obtained and 6 of them (A1, A3, A4, A5, A9, A10)
were sequenced. The sequences are similar to Leuconostoc mesenteroides
(GenBank Access No. AY453065), some uncultured bacteria (AJ318147,
AF227834, AJ576427), Ethanologenbacterium (AY434722), Clostridiaceae
(AB084627), etc. In order to investigate the SRBs in the reactor,the
active sludge was cultured on SRB-selected media and also did SSCP
with the compound cultured bacteria. Two new bands appeared, one
similar to Bacteroidetes (AB074606) and another similar to
Desulfovibrio (Y12254, U42221). The experimental results indicate that
the proportion of SRBs in the reactor is probably less than 1.5
percent. But the few SRBs play a very important role during the course
of sulfate reduction via cooperation with acidogenic.
PMID: 16212191 [PubMed - in process]
6: Mycol Res. 2005 Sep;109(Pt 9):1015-28.
Specific and sensitive PCR-based detection of Septoria musiva, S.
populicola and S. populi, the causes of leaf spot and stem canker on
poplars.
Feau N, Weiland JE, Stanosz GR, Bernier L.
Centre de Recherche en Biologie Forestiere, Universite Laval, SainteFoy, Quebec G1K 7P4, Canada.
The development of a PCR assay for the detection of the poplar
pathogenic fungi Septoria musiva (teleomorph Mycosphaerella populorum),
S. populicola (M. populicola) and S. populi (M. populi) is described.
Three pairs of species-specific PCR primers were designed using
interspecific polymorphisms in the internal transcribed spacer (ITS) of
nuclear ribosomal RNA gene (rDNA) repeats. The specificity of the three
primer pairs was successfully tested on a collection of 40 S. musiva,
39 S. populicola and six S. populi isolates. Using stringent PCR
conditions, no cross-reaction was observed with any of the isolates
tested. The specificity of the PCR assay was further confirmed with DNA
extracted from 12 additional Septoria species and 17 other fungal
species obtained from stems or leaves of poplars. Specific
amplification of the fragments for S. musiva and S. populicola was
sensitive relatively to the technique used, detecting as low as 1 pg
template DNA, and 10 pg of DNA of the target species in a background of
1 ng of DNA of the other species. Moreover, using DNA purified directly
from disrupted conidia, it was possible to detect with a probability of
90%, using one unique PCR assay, the DNA equivalent of 166 conidia per
microl of S. musiva and 156 conidia per microl of S. populicola. The
procedures developed in this work can thus be applied for rapid and
accurate detection and identification of Septoria species from poplars.
PMID: 16209307 [PubMed - in process]
7: J Neurosci. 2005 Oct 5;25(40):9171-5.
Ribosome dysfunction is an early event in Alzheimer's disease.
Ding Q, Markesbery WR, Chen Q, Li F, Keller JN.
Department of Anatomy and Neurobiology, University of Kentucky,
Lexington, Kentucky 40536-0230, USA.
Alzheimer's disease (AD) is a progressive and devastating disorder that
is often preceded by mild cognitive impairment (MCI). In the present
study, we report that in multiple cortical areas of MCI and AD
subjects, there is a significant impairment in ribosome function that
is not observed in the cerebellum of the same subjects. The impairment
in ribosome function is associated with a decreased rate and capacity
for protein synthesis, decreased ribosomal RNA and tRNA levels, and
increased RNA oxidation. No alteration in the level of initiation
factors was observed in the brain regions exhibiting impairments in
protein synthesis. Together, these data indicate for the first time
that impairments in protein synthesis may be one of the earliest
neurochemical alterations in AD and directly demonstrate that the
polyribosome complex is adversely affected early in the development of
AD. These data have important implications for AD studies involving
proteomics and studies analyzing proteolysis in AD, indicate that
oxidative damage may contribute to decreased protein synthesis, and
suggest a role for alterations in protein synthesis as a novel
contributor to the onset and development of AD.
PMID: 16207876 [PubMed - in process]
8: Int J Hematol. 2005 Oct;82(3):184-9.
Dyskeratosis congenita: a disorder of defective telomere maintenance?
Walne AJ, Marrone A, Dokal I.
Department of Haematology, Division of Investigative Science, Faculty
of Medicine, Imperial College London, Hammersmith Hospital, London,
United Kingdom.
Dyskeratosis congenita (DC) is a rare multisystem bone marrow failure
syndrome that displays marked clinical and genetic heterogeneity. Xlinked recessive, autosomal dominant and autosomal recessive forms of
the disease are recognized. The gene that is mutated in the X-linked
form of the disease is DKC1. The DKC1-encoded protein, dyskerin, is a
component of small nucleolar ribonucleoprotein particles, which are
important in ribosomal RNA processing, and of the telomerase complex.
The autosomal dominant form of DC is due to mutations in the gene for
the RNA component of telomerase (TERC). Because both dyskerin and TERC
are components of the telomerase complex and all patients with DC have
short telomeres, the principal pathology of DC appears to relate to
telomerase dysfunction, although defects in ribosomal processing via
dyskerin's involvement in pseudouridylation cannot be completely ruled
out. The gene or genes involved in autosomal recessive DC remain
elusive, although genes whose products are required for telomere
maintenance remain strong candidates. The study of DC highlights the
importance of telomerase in humans and how its deficiency results in
multiple abnormalities, including premature aging, bone marrow failure,
and cancer.
PMID: 16207588 [PubMed - in process]
9: J Cell Physiol. 2005 Oct 4; [Epub ahead of print]
Ki-67 protein is associated with ribosomal RNA transcription in
quiescent and proliferating cells.
Bullwinkel J, Baron-Luhr B, Ludemann A, Wohlenberg C, Gerdes J,
Scholzen T.
Division of Tumour Biology, Research Center Borstel, Borstel, Germany.
The nuclear Ki-67 protein (pKi-67) has previously been shown to be
exclusively expressed in proliferating cells. As a result, antibodies
against this protein are widely used as prognostic tools in cancer
diagnostics. Here we show, that despite the strong downregulation of
pKi-67 expression in non-proliferating cells, the protein can
nevertheless be detected at sites linked to ribosomal RNA (rRNA)
synthesis. Although this finding does not argue against the use of pKi67 as a proliferation marker, it has wide ranging implications for the
elucidation of pKi-67 function. Employing the novel antibody TuBB-9, we
could further demonstrate that also in proliferating cells, a fraction
of pKi-67 is found at sites linked to the rRNA transcription machinery
during interphase and mitosis. Moreover, chromatin immunoprecipitation
(ChIP) assays provided evidence for a physical association of pKi-67
with chromatin of the promoter and transcribed region of the rRNA gene
cluster. These data strongly suggest a role for pKi-67 in the early
steps of rRNA synthesis. (c) 2005 Wiley-Liss, Inc.
PMID: 16206250 [PubMed - as supplied by publisher]
10: Mar Biotechnol (NY). 2005 Sep 28; [Epub ahead of print]
Mitochondrial DNA Sequence and Gene Organization in Australian Backup
Abalone Haliotis Rubra (Leach).
Maynard BT, Kerr LJ, McKiernan JM, Jansen ES, Hanna PJ.
School of Biological & Chemical Sciences, Deakin University, Geelong,
VIC 3217, Australia, pjh@deakin.edu.au.
The complete mitochondrial DNA of the blacklip abalone Haliotis rubra
(Gastropoda: Mollusca) was cloned and 16,907 base pairs were sequenced.
The sequence represents an estimated 99.85% of the mitochondrial
genome, and contains 2 ribosomal RNA, 22 transfer RNA, and 13 proteincoding genes found in other metazoan mtDNA. An AT tandem repeat and a
possible C-rich domain within the putative control region could not be
fully sequenced. The H. rubra mtDNA gene order is novel for mollusks,
separated from the black chiton Katharina tunicata by the individual
translocations of 3 tRNAs. Compared with other mtDNA regions, sequences
from the ATP8, NAD2, NAD4L, NAD6, and 12S rRNA genes, as well as the
control region, are the most variable among representatives from
Mollusca, Arthropoda, and Rhynchonelliformea, with similar mtDNA
arrangements to H. rubra. These sequences are being evaluated as
genetic markers within commercially important Haliotis species, and
some applications and considerations for their use are discussed.
PMID: 16206015 [PubMed - as supplied by publisher]
11: Cell Cycle. 2005 Aug;4(8):1036-8. Epub 2005 Aug 20.
Cellular stress and nucleolar function.
Mayer C, Grummt I.
Division of Molecular Biology of the Cell II, German Cancer Research
Center, Heidelberg, Germany.
All organisms sense and respond to conditions that stress their
homeostatic mechanisms. Here we review current studies showing that the
nucleolus, long regarded as a mere ribosome producing factory, plays a
key role in monitoring and responding to cellular stress. After
exposure to extra- or intracellular stress, cells rapidly down-regulate
the synthesis of ribosomal RNA. Impairment of nucleolar function in
response to stress is accompanied by perturbation of nucleolar
structure, cell cycle arrest and stabilization of p53. The nucleolar
target for down-regulation of rDNA transcription is TIF-IA, an
essential transcription factor that modulates the activity of RNA
polymerase I (Pol I). Upon stress, TIF-IA is phosphorylated by c-Jun Nterminal kinase 2 (JNK2). Phosphorylation prevents TIF-IA from
interaction with Pol I, thereby impairing transcription complex
formation and rRNA synthesis. Furthermore, stress-induced inactivation
of TIF-IA is accompanied by translocation of TIF-IA from the nucleolus
to the nucleoplasm. These findings, together with other data showing
stress-induced release of nucleolar proteins to carry out other
regulatory functions, reinforce the growing realization that nucleoli
orchestrate the chain of events the cell uses to properly respond to
stress signals.
PMID: 16205120 [PubMed - in process]
12: Mol Ecol. 2005 Oct;14(12):3787-800.
Genetic divergence does not predict change in ornament expression among
populations of stalk-eyed flies.
Swallow JG, Wallace LE, Christianson SJ, Johns PM, Wilkinson GS.
Department of Biology, University of South Dakota, Vermillion, SD
57069, USA.
Stalk-eyed flies (Diptera: Diopsidae) possess eyes at the ends of
elongated peduncles, and exhibit dramatic variation in eye span,
relative to body length, among species. In some sexually dimorphic
species, evidence indicates that eye span is under both intra- and
intersexual selection. Theory predicts that isolated populations should
evolve differences in sexually selected traits due to drift. To
determine if eye span changes as a function of divergence time,
1370 flies from 10 populations of the sexually dimorphic species,
Cyrtodiopsis dalmanni and Cyrtodiopsis whitei, and one population of
the sexually monomorphic congener, Cyrtodiopsis quinqueguttata, were
collected from Southeast Asia and measured. Genetic differentiation was
used to assess divergence time by comparing mitochondrial (cytochrome
oxidase II and 16S ribosomal RNA gene fragments) and nuclear (wingless
gene fragment) DNA sequences for c. five individuals per population.
Phylogenetic analyses indicate that most populations cluster as
monophyletic units with up to 9% nucleotide substitutions between
populations within a species. Analyses of molecular variance suggest a
high degree of genetic structure within and among the populations; >
97% of the genetic variance occurs between populations and species
while < 3% is distributed within populations, indicating that most
populations have been isolated for thousands of years. Nevertheless,
significant change in the allometric slope of male eye span on body
length was detected for only one population of either dimorphic
species. These results are not consistent with genetic drift. Rather,
relative eye span appears to be under net stabilizing selection in most
populations of stalk-eyed flies. Given that one population exhibited
dramatic evolutionary change, selection, rather than genetic variation,
appears to constrain eye span evolution.
PMID: 16202096 [PubMed - in process]
13: RNA. 2005 Oct;11(10):1478-84.
Are stop codons recognized by base triplets in the large ribosomal RNA
subunit?
Liang H, Landweber LF, Fresco JR.
Department of Molecular Biology, Princeton University, Princeton, NJ
08544, USA.
The precise mechanism of stop codon recognition in translation
termination is still unclear. A previously published study by Ivanov
and colleagues proposed a new model for stop codon recognition in which
3-nucleotide Ter-anticodons within the loops of hairpin helices 69
(domain IV) and 89 (domain V) in large ribosomal subunit (LSU) rRNA
recognize stop codons to terminate protein translation in eubacteria
and certain organelles. We evaluated this model by extensive
bioinformatic analysis of stop codons and their putative corresponding
Ter-anticodons across a much wider range of species, and found many
cases for which it cannot explain the stop codon usage without
requiring the involvement of one or more of the eight possible
noncomplementary base pairs. Involvement of such base pairs may not be
structurally or thermodynamically damaging to the model. However, if,
according to the model, Ter-anticodon interaction with stop codons
occurs within the ribosomal A-site, the structural stringency which
that site imposes on sense codon.tRNA anticodon interaction should also
extend to stop codon.Ter-anticodon interactions. Moreover, with TertRNA in place of an aminoacyl-tRNA, for each of the various Teranticodons there is a sense codon that can interact with it
preferentially by complementary and wobble base-pairing. Both these
considerations considerably weaken the arguments put forth previously.
PMID: 16199759 [PubMed - in process]
14: Biosci Biotechnol Biochem. 2005 Sep;69(9):1793-7.
Culture-independent analysis of fecal microbiota in cattle.
Ozutsumi Y, Hayashi H, Sakamoto M, Itabashi H, Benno Y.
United Graduate School of Agricultural Science, Tokyo University of
Agriculture and Technology.
The phylogenetic diversity of the fecal bacterial community in Holstein
cattle was determined by 16S ribosomal RNA gene sequence analysis. The
sequences were affiliated with the following phyla: Firmicutes (81.3%),
Bacteroidetes (14.4%), Actinobacteria (2.5%), and Proteobacteria
(1.4%). The Clostridium leptum subgroup was the most phylogenetically
diverse group in cattle feces. In addition, a number of previously
uncharacterized and unidentified bacteria were recognized in clone
libraries.
PMID: 16195605 [PubMed - in process]
15: Q Rev Biophys. 2004 Nov;37(3-4):197-284.
Translation initiation: structures, mechanisms and evolution.
Marintchev A, Wagner G.
Department of Biological Chemistry and Molecular Pharmacology, Harvard
Medical School, Boston, USA.
Translation, the process of mRNA-encoded protein synthesis, requires a
complex apparatus, composed of the ribosome, tRNAs and additional
protein factors, including aminoacyl tRNA synthetases. The ribosome
provides the platform for proper assembly of mRNA, tRNAs and protein
factors and carries the peptidyl-transferase activity. It consists of
small and large subunits. The ribosomes are ribonucleoprotein particles
with a ribosomal RNA core, to which multiple ribosomal proteins are
bound. The sequence and structure of ribosomal RNAs, tRNAs, some of the
ribosomal proteins and some of the additional protein factors are
conserved in all kingdoms, underlying the common origin of the
translation apparatus. Translation can be subdivided into several
steps: initiation, elongation, termination and recycling. Of these,
initiation is the most complex and the most divergent among the
different kingdoms of life. A great amount of new structural,
biochemical and genetic information on translation initiation has been
accumulated in recent years, which led to the realization that
initiation also shows a great degree of conservation throughout
evolution. In this review, we summarize the available structural and
functional data on translation initiation in the context of evolution,
drawing parallels between eubacteria, archaea, and eukaryotes. We will
start with an overview of the ribosome structure and of translation in
general, placing emphasis on factors and processes with relevance to
initiation. The major steps in initiation and the factors involved will
be described, followed by discussion of the structure and function of
the individual initiation factors throughout evolution. We will
conclude with a summary of the available information on the
kinetic and thermodynamic aspects of translation initiation.
PMID: 16194295 [PubMed - in process]
16: EMBO J. 2005 Sep 29; [Epub ahead of print]
Structure of a Mycobacterium tuberculosis NusA-RNA complex.
Beuth B, Pennell S, Arnvig KB, Martin SR, Taylor IA.
Division of Protein Structure, National Institute for Medical Research,
London, UK.
NusA is a key regulator of bacterial transcriptional elongation,
pausing, termination and antitermination, yet relatively little is
known about the molecular basis of its activity in these fundamental
processes. In Mycobacterium tuberculosis, NusA has been shown to bind
with high affinity and specificity to BoxB-BoxA-BoxC antitermination
sequences within the leader region of the single ribosomal RNA (rRNA)
operon. We have determined high-resolution X-ray structures of a
complex of NusA with two short oligo-ribonucleotides derived from the
BoxC stem-loop motif and have characterised the interaction of NusA
with a variety of RNAs derived from the antitermination region. These
structures reveal the RNA bound in an extended conformation to a large
interacting surface on both KH domains. Combining structural data with
observed spectral and calorimetric changes, we now show that NusA
binding destabilises secondary structure within rRNA antitermination
sequences and propose a model where NusA functions as a chaperone for
nascently forming RNA structures.
PMID: 16193062 [PubMed - as supplied by publisher]
17: Proc Biol Sci. 2005 Oct 7;272(1576):2073-81.
The extent of protist diversity: insights from molecular ecology of
freshwater eukaryotes.
Slapeta J, Moreira D, Lopez-Garcia P.
Systematique et Evolution, UMR CNRS 8079, Universite Paris-Sud Unite
d'Ecologie 91405 Orsay Cedex, France.
Classical studies on protist diversity of freshwater environments
worldwide have led to the idea that most species of microbial
eukaryotes are known. One exemplary case would be constituted by the
ciliates, which have been claimed to encompass a few thousands of
ubiquitous species, most of them already described. Recently, molecular
methods have revealed an unsuspected protist diversity, especially in
oceanic as well as some extreme environments, suggesting the
occurrence of a hidden diversity of eukaryotic lineages. In order to
test if this holds also for freshwater environments, we have carried
out a molecular survey of small subunit ribosomal RNA genes in water
and sediment samples of two ponds, one oxic and another suboxic, from
the same geographic area. Our results show that protist diversity is
very high. The majority of phylotypes affiliated within a few well
established eukaryotic kingdoms or phyla, including alveolates,
cryptophytes, heterokonts, Cercozoa, Centroheliozoa and haptophytes,
although a few sequences did not display a clear taxonomic affiliation.
The diversity of sequences within groups was very large, particularly
that of ciliates, and a number of them were very divergent from known
species, which could define new intra-phylum groups. This suggests
that, contrary to current ideas, the diversity of freshwater protists
is far from being completey described.
PMID: 16191619 [PubMed - in process]
18: Histochem Cell Biol. 2005 Sep 27;:1-10 [Epub ahead of print]
Re-localization of nuclear DNA helicase II during the growth period of
bovine oocytes.
Baran V, Kovarova H, Klima J, Hozak P, Motlik J.
Institute of Animal Physiology, Slovak Academy of Sciences, Kosice,
Slovakia.
Nuclear DNA helicase II (NDH II) is the bovine homolog of human RNA
helicase A. The aim of this study was to compare NDH II localization
between somatic cells (bovine embryonal fibroblasts) and female germ
cells (oocytes), with the main focus on the dynamic changes in the
redistribution of NDH II during the growth phase of the bovine oocytes.
The fine granular staining of NDH II was spread in the whole
nucleoplasm of fibroblasts, excluding the reticulated nucleoli. In
contrast, the large reticulated nucleoli of the growing oocytes
isolated from early antral follicles exhibited strong positivity for
NDH II together with the immunostaining signals of upstream binding
factor (UBF) and RNA polymerase I subunit (PAF53), documenting the high
synthetic activity of these nucleoli. At the time of termination of
oocyte growth, NDH II was preferentially located at the nucleolar
periphery together with proteins of fibrillar centres. In fully
grown oocytes, NDH II was still present in the thin periphery shell
around the compact nucleolar core. The semiquantitative RT-PCR revealed
that the average signal of NDH II mRNA in fully grown oocytes was only
at 40% level in comparison with growing oocytes. Western blot analysis
further confirmed that a 140 kD NDH II protein was abundant in growing
oocytes, while the signal was substantially weaker in fully grown
oocytes. The significant decrease in NDH II gene expression and in NDH
II mRNA translation correlates with a termination of the oocyte growth.
Altogether, the results demonstrate that NDH II expression parallels
the activity of ribosomal RNA biosynthesis in the bovine growing
oocytes.
PMID: 16187064 [PubMed - as supplied by publisher]
19: J Biol Chem. 2005 Sep 26; [Epub ahead of print]
Microspherule protein 1, Mi-2beta, and ret finger protein associate in
the nucleolus and up-regulate ribosomal gene transcription.
Shimono K, Shimono Y, Shimokata K, Ishiguro N, Takahashi M.
Department of Pathology, Nagoya University, Nagoya 466-8550.
The nucleolus is the site of ribosomal DNA (rDNA) transcription and
ribosome production. In exploring the role of nucleolar protein MCRS1
(microspherule protein1)/MSP58 (58 kDa microspherule protein), we found
that Mi-2beta, a component of a nucleosome remodeling and deacetylase
(NuRD) complex, RET finger protein (RFP), and upstream binding factor
(UBF) were associated with MCRS1. Yeast two-hybrid assays revealed that
MCRS1 bound to the ATPase/helicase region of Mi-2beta and the coiledcoil region of RFP. Interestingly, confocal microscopic analyses
revealed the co-localization of MCRS1, Mi-2beta, RFP, and the rRNA
transcription factor UBF in the nucleoli. We also found that MCRS1,
Mi-2beta, and RFP were associated with rDNA using a chromatin
immunoprecipitation assay. Finally, we showed that MCRS1, Mi-2beta, and
RFP up-regulated transcriptional activity of the rDNA promoter, and
that ribosomal RNA transcription was repressed when MCRS1, Mi-2beta,
and RFP expression was reduced using siRNA. These results indicated
that Mi-2beta and RFP, known to be involved in transcriptional
repression in the nucleus, co-localize with MCRS1 in the nucleolus and
appear to activate the rRNA transcription.
PMID: 16186106 [PubMed - as supplied by publisher]
20: BMC Genomics. 2005 Sep 26;6:136.
Mitochondrial-encoded membrane protein transcripts are pyrimidine-rich
while soluble protein transcripts and ribosomal RNA are purine-rich.
Bradshaw PC, Rathi A, Samuels DC.
Virginia Bioinformatics Institute, Virginia Polytechnic Institute and
State University, Blacksburg, VA 24061, USA. patrickbradshaw@yahoo.com
BACKGROUND: Eukaryotic organisms contain mitochondria, organelles
capable of producing large amounts of ATP by oxidative phosphorylation.
Each cell contains many mitochondria with many copies of mitochondrial
DNA in each organelle. The mitochondrial DNA encodes a small but
functionally critical portion of the oxidative phosphorylation
machinery, a few other species-specific proteins, and the rRNA and tRNA
used for the translation of these transcripts. Because the
microenvironment of the mitochondrion is unique, mitochondrial genes
may be subject to different selectional pressures than those affecting
nuclear genes. RESULTS: From an analysis of the mitochondrial genomes
of a wide range of eukaryotic species we show that there are three
simple rules for the pyrimidine and purine abundances in mitochondrial
DNA transcripts. Mitochondrial membrane protein transcripts are
pyrimidine rich, rRNA transcripts are purine-rich and the soluble
protein transcripts are purine-rich. The transitions between
pyrimidine and purine-rich regions of the genomes are rapid and are
easily visible on a pyrimidine-purine walk graph. These rules are
followed, with few exceptions, independent of which strand encodes the
gene. Despite the robustness of these rules across a diverse set of
species, the magnitude of the differences between the pyrimidine and
purine content is fairly small. Typically, the mitochondrial membrane
protein transcripts have a pyrimidine richness of 56%, the rRNA
transcripts are 55% purine, and the soluble protein transcripts are
only 53% purine. CONCLUSION: The pyrimidine richness of mitochondrialencoded membrane protein transcripts is partly driven by U nucleotides
in the second codon position in all species, which yields hydrophobic
amino acids. The purine-richness of soluble protein transcripts is
mainly driven by A nucleotides in the first codon position. The purinerichness of rRNA is also due to an abundance of A nucleotides. Possible
mechanisms as to how these trends are maintained in mtDNA genomes of
such diverse ancestry, size and variability of A-T richness are
discussed.
PMID: 16185363 [PubMed - in process]
21: Gene. 2005 Nov 7;360(2):92-102. Epub 2005 Sep 23.
The mitochondrial genome sequence of the scorpion Centruroides limpidus
(Karsch 1879) (Chelicerata; Arachnida).
Davila S, Pinero D, Bustos P, Cevallos MA, Davila G.
Programa de Genomica Evolutiva, Centro de Ciencias Genomicas - UNAM,
AvenidaUniversidad, s/n, Chamilpa, Cuernavaca 62210, Apartado Postal
565-A, Morelos, Mexico.
The mitochondrial genome of the scorpion Centruroides limpidus
(Chelicerata; Arachnida) has been completely sequenced and is 14519 bp
long. The genome contains 13 protein-encoding genes, two ribosomal RNA
genes, 21 transfer RNA genes and a large non-coding region related to
the control region. The overall A+T composition is the lowest among the
complete mitochondrial sequences published within the Chelicerata
subphylum. Gene order and gene content differ slightly from that of
Limulus polyphemus (Chelicerata: Xiphosura): i.e., the lack of the trnD
gene, and the translocation-inversion of the trnI gene. Preliminary
phylogenetic analysis of some Chelicerata shows that scorpions (C.
limpidus and Mesobuthus gibbosus) make a tight cluster with the spiders
(Arachnida; Araneae). Our analysis does not support that Scorpiones
order is the sister group to all Arachnida Class, since it is closer to
Araneae than to Acari orders.
PMID: 16183216 [PubMed - in process]
22: Mol Biochem Parasitol. 2005 Sep 20; [Epub ahead of print]
Tagging a T. brucei RRNA locus improves stable transfection efficiency
and circumvents inducible expression position effects.
Alsford S, Kawahara T, Glover L, Horn D.
London School of Hygiene & Tropical Medicine, Department of Infections
and Tropical Diseases, Keppel Street, London WC1E 7HT, UK.
In Trypanosoma brucei, RNA interference (RNAi) and recombinant protein
expression are established as powerful approaches for functional
genomics, particularly when combined with inducible expression. The
favoured methods involve exploiting homologous recombination to target
expression cassettes to a chromosome sub-set to establish stable cell
lines. Unfortunately, bloodstream-form cells, those that cause disease
in mammals, exhibit low efficiency stable transfection. Current
expression systems can also exhibit other undesirable features,
including variable position effects and leaky, inducible expression. We
have developed systems in bloodstream-form cells that alleviate these
problems. Using constructs for RNAi and expression of (GFP) tagged
proteins, we target a (hyg) tagged ribosomal RNA (RRNA) locus which
circumvents position effects and allows increased targeting efficiency.
We also report a compatible double-inducible system for tight
regulation of highly toxic products. This system exploits a new
inducible RRNA promoter to drive T7 RNA polymerase (T7RNAP)
transcription which then drives expression from inducible T7
promoters. The developments described should facilitate functional
analysis and increased throughput.
PMID: 16182389 [PubMed - as supplied by publisher]
23: J Biomed Opt. 2005 Jul-Aug;10(4):44025.
Direct visualization of mRNA colocalization with mitochondria in living
cells using molecular beacons.
Santangelo PJ, Nitin N, Bao G.
Georgia Institute of Technology and Emory University, Department of
Biomedical Engineering, Atlanta, Georgia 30332.
The intracellular localization and specific organelle association of
mRNA may reflect essential functions, stages, and stability of mRNA. We
report the direct visualization of subcellular localization of K-ras
and glyceraldehydes 3-phosphate dehydrogenase (GAPDH) mRNAs in live HDF
cells using molecular beacons together with membrane-permeabilization
and peptide-based delivery. Unexpectedly, we found that both K-ras and
GAPDH mRNAs colocalize with mitochondria. Extensive control studies are
performed, including the use of fluorescence in-situ hybridization
(FISH), negative-control beacons, and the detection of colocalization
of 28S ribosomal RNA with the rough endoplasmic reticulum (ER),
suggesting that the mRNA localization and colocalization patterns
observed in our study are true and specific. Our observation reveals
intriguing subcellular associations of mRNA with organelles such as
mitochondria, which may provide new insight into the transport,
dynamics, and functions of mRNA and mRNA-protein interactions.
PMID: 16178658 [PubMed - in process]
24: Parasitology. 2005 Sep;131(Pt 3):321-9.
Differential polyadenylation of ribosomal RNA during posttranscriptional processing in Leishmania.
Decuypere S, Vandesompele J, Yardley V, De Donckeri S, Laurent T, Rijal
S, Llanos-Cuentas A, Chappuis F, Arevalo J, Dujardin JC.
Department of Parasitology, Unit of Molecular Parasitology, Prince
Leopold Institute of Tropical Medicine, Antwerp B-2000, Belgium.
The protozoan parasite Leishmania belongs to the most ancient
eukaryotic lineages and this is reflected in several distinctive
biological features, such as eukaryotic polycistronic transcription and
RNA trans-splicing. The disclosure of this organism's unusual
characteristics leads to a better understanding of the origin and
nature of fundamental biological processes in eukaryotes. Here we
report another unusual phenomenon as we demonstrate that precursor
ribosomal RNA can be extensively polyadenylated during posttranscriptional processingt. Furthermore, we demonstrate that the
degree of precursor rRNA polyadenylation is variable in different
strains and in the different life-stages of a strain.
PMID: 16178353 [PubMed - in process]
25: Nature. 2005 Sep 22;437(7058):543-6.
Isolation of an autotrophic ammonia-oxidizing marine archaeon.
Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl
DA.
Department of Civil and Environmental Engineering, University of
Washington, Seattle, Washington 98195, USA.
For years, microbiologists characterized the Archaea as obligate
extremophiles that thrive in environments too harsh for other
organisms. The limited physiological diversity among cultivated Archaea
suggested that these organisms were metabolically constrained to a few
environmental niches. For instance, all Crenarchaeota that are
currently cultivated are sulphur-metabolizing thermophiles. However,
landmark studies using cultivation-independent methods uncovered vast
numbers of Crenarchaeota in cold oxic ocean waters. Subsequent
molecular surveys demonstrated the ubiquity of these low-temperature
Crenarchaeota in aquatic and terrestrial environments. The numerical
dominance of marine Crenarchaeota--estimated at 10(28) cells in the
world's oceans--suggests that they have a major role in global
biogeochemical cycles. Indeed, isotopic analyses of marine crenarchaeal
lipids suggest that these planktonic Archaea fix inorganic carbon. Here
we report the isolation of a marine crenarchaeote that grows
chemolithoautotrophically by aerobically oxidizing ammonia to nitrite-the first observation of nitrification in the Archaea. The autotrophic
metabolism of this isolate, and its close phylogenetic relationship to
environmental marine crenarchaeal sequences, suggests that nitrifying
marine Crenarchaeota may be important to global carbon and nitrogen
cycles.
PMID: 16177789 [PubMed - indexed for MEDLINE]
26: Dis Aquat Organ. 2005 Aug 9;66(1):1-7.
Spinal curvature of cultured Japanese mackerel Scomber japonicus
associated with a brain myxosporean, Myxobolus acanthogobii.
Yokoyama H, Freeman MA, Itoh N, Fukuda Y.
Department of Aquatic Bioscience, Graduate School of Agricultural and
Life Sciences, The University of Tokyo, Bunkyo, Tokyo 113-8657, Japan.
ayokoh@mail.ecc.u-tokyo.ac.jp
Skeletal deformities were found in the cultured Japanese mackerel
Scomber japonicus. External and radiographical observations showed the
deformed fish to exhibit a dorso-ventral spinal curvature (kyphosis)
without fracture or dislocation of the vertebrae. Numerous myxosporean
cysts, ca. 0.3 to 1.0 mm in diameter, formed in the 4th ventricle, the
cavity of the optic tectum, the surface of the olfactory lobe and bulb,
the optic lobe and the inferior lobe of the brain. Spore morphology and
molecular analysis of the small subunit ribosomal RNA gene sequence
identified the myxosporean parasite as Myxobolus acanthogobii, a
parasite which also causes scoliosis in yellowtail Seriola
quinqeradiata. Histopathological observation showed that the
myxosporean cysts were encapsulated within the host's collagenous layer
although some had disintegrated to disperse mature spores into the
cranial cavity. Occasionally, lymphocytic infiltration and local
granulomatous inflammation were found to be associated with spore
dispersion.
PMID: 16175961 [PubMed - in process]
27: Adv Space Res. 2005;35(9):1634-42.
Analysis of the archaeal sub-seafloor community at Suiyo Seamount on
the Izu-Bonin Arc.
Hara K, Kakegawa T, Yamashiro K, Maruyama A, Ishibashi J, Marumo K,
Urabe T, Yamagishi A.
Department of Molecular Biology, Tokyo University of Pharmacy and Life
Science, Hachioji, Tokyo, Japan.
A sub-surface archaeal community at the Suiyo Seamount in the Western
Pacific Ocean was investigated by 16S rRNA gene sequence and whole-cell
in situ hybridization analyses. In this study, we drilled and cased
holes at the hydrothermal area of the seamount to minimize
contamination of the hydrothermal fluid in the sub-seafloor by
penetrating seawater. PCR clone analysis of the hydrothermal fluid
samples collected from a cased hole indicated the presence of
chemolithoautotrophic primary biomass producers of Archaeoglobales and
the Methanococcales-related archaeal HTE1 group, both of which can
utilize hydrogen as an electron donor. We discuss the implication of
the microbial community on the early history of life and on the search
for extraterrestrial life. c2005 COSPAR. Published by Elsevier Ltd. All
rights reserved.
PMID: 16175703 [PubMed - indexed for MEDLINE]
28: Genetica. 2005 Sep;125(1):27-32.
Comparative molecular cytogenetic analysis of two congeneric species,
mugil curema and m. Liza (pisces, mugiliformes), characterized by
significant karyotype diversity.
Rossi AR, Gornung E, Sola L, Nirchio M.
Department of Animal and Human Biology, University of Rome I "La
Sapienza", via A. Borelli, 50, 00161, Rome, Italy,
luciana.sola@uniroma1.it.
Two congeneric mullet species, Mugil liza and M. curema, respectively
with an all-uniarmed and an all-biarmed karyotype, were cytogenetically
studied by base-specific fluorochrome staining and FISH-mapping of 45S
and 5S ribosomal RNA genes (rDNA) and the (TTAGGG)(n) telomeric
repeats. Whereas 45S rDNA sites might be homeologus in the two species,
5S rDNA sites are not, as they are localized on chromosome arms of
different size. In both species, the (TTAGGG)(n) telomeric probe
hybridized to natural telomeres and was found scattered along the NORs.
In metacentric chromosomes of M. curema, no pericentromeric signals of
the telomeric probe were detected. Data are discussed in relation to
the karyotype evolution in Mugilidae and to the mechanisms and the
evolutionary implications of Robertsonian rearrangements in M. curema.
PMID: 16175452 [PubMed - in process]
29: Am J Vet Res. 2005 Aug;66(8):1380-5.
Evaluation of a multiplex polymerase chain reaction assay for
simultaneous detection of Rhodococcus equi and the vapA gene.
Halbert ND, Reitzel RA, Martens RJ, Cohen ND.
Department of Large Animal Clinical Sciences, College of Veterinary
Medicine and Biomedical Sciences, Texas A&M University, College
Station, TX 77843-4475, USA.
OBJECTIVE: To evaluate sensitivity and specificity of a multiplex
polymerase chain reaction (PCR) assay for simultaneous detection of
Rhodococcus equi and differentiation of strains that contain the
virulence-associated gene (vapA) from strains that do not. SAMPLE
POPULATION: 187 isolates of R equi from equine and nonequine tissue and
environmental specimens and 27 isolates of bacterial species
genetically or morphologically similar to R equi. PROCEDURE: The
multiplex PCR assay included 3 gene targets: a universal 311-bp
bacterial 16S ribosomal RNA amplicon (positive internal control), a
959-bp R equi-specific target in the cholesterol oxidase gene (choE),
and a 564-bp amplicon of the vapA gene. Duplicate multiplex PCR assays
for these targets and confirmatory singleplex PCR assays for vapA and
choE were performed for each R equi isolate. An additional PCR assay
was used to examine isolates for the vapB gene. RESULTS: Results of
duplicate multiplex and singleplex PCR assays were correlated in all
instances, revealing high specificity and reliability (reproducibility)
of the vapA multiplex assay. Of the pulmonary isolates from horses with
suspected R equi pneumonia, 97.4% (76/78) yielded positive results for
vapA. Seven of 50 (14%) human isolates of R equi yielded positive
results for vapA. Six human R equi isolates and 1 porcine isolate
yielded positive results for vapB. No isolates with vapA and vapB genes
were detected. CONCLUSIONS AND CLINICAL RELEVANCE: The multiplex PCR
assay is a sensitive and specific method for simultaneous confirmation
of species identity and detection of the vapA gene. The assay appeared
to be a useful tool for microbiologic and epidemiologic diagnosis and
research.
Publication Types:
Evaluation Studies
PMID: 16173481 [PubMed - indexed for MEDLINE]
30: Arch Otolaryngol Head Neck Surg. 2005 Sep;131(9):804-8.
Prevalence of Helicobacter pylori DNA in recurrent aphthous ulcerations
in mucosa-associated lymphoid tissues of the pharynx.
Elsheikh MN, Mahfouz ME.
Department of Otolaryngology, Tanta University, Tanta, Egypt.
mnel_sheikh@hotmail.com
OBJECTIVE: To determine the presence of Helicobacter pylori and, if
detected, its potential prevalence in causing recurrent aphthous ulcers
confined to mucosa-associated lymphoid tissues of the pharynx. DESIGN:
Prospective, controlled clinical trial. SETTING: Otolaryngology
Department of Tanta University Hospitals, Tanta, Egypt. PATIENTS: A
total of 146 patients with recurrent multiple aphthous ulcers of the
oral cavity and pharynx and 20 normal control subjects. INTERVENTIONS:
Patients were assigned to group 1 (n = 58), in which the ulcers were
strictly limited to the lymphoid tissues, or group 2 (n = 88), in which
the ulcers were randomly distributed in the oral cavity and pharynx.
Helicobacter pylori DNA was extracted from 3-mm-diameter tissue
samples, and polymerase chain reaction amplifications were performed
for the 16S ribosomal RNA gene. MAIN OUTCOME MEASURE: Positivity for H
pylori. RESULTS: In group 1, 39 patients (67%) were positive for H
pylori DNA, while in group 2, 9 patients (10%) were positive (chi(2)
test, P<.001). It was not detected in any of the 20 control samples.
CONCLUSION: Our results support a possible causative role for H pylori
in recurrent aphthous ulcerations with a characteristic distribution
and affinity to mucosa-associated lymphoid tissues of the pharynx.
PMID: 16172360 [PubMed - indexed for MEDLINE]
31: Protist. 2005 Aug;156(2):215-24.
18S ribosomal RNA gene sequences of Cochliopodium (Himatismenida) and
the phylogeny of Amoebozoa.
Kudryavtsev A, Bernhard D, Schlegel M, Chao EE, Cavalier-Smith T.
Department of Invertebrate Zoology, Faculty of Biology and Soil
Science, Saint-Petersburg State University, Saint-Petersburg, 199034,
Russia. aak@ak14261.spb.edu
Cochliopodium is a very distinctive genus of discoid amoebae covered by
a dorsal tectum of carbohydrate microscales. Its phylogenetic position
is unclear, since although sharing many features with naked
"gymnamoebae", the tectum sets it apart. We sequenced 18S ribosomal RNA
genes from three Cochliopodium species (minus, spiniferum and
Cochliopodium sp., a new species resembling C. minutum). Phylogenetic
analysis shows Cochliopodium as robustly holophyletic and within
Amoebozoa, in full accord with morphological data. Cochliopodium is
always one of the basal branches within Amoebozoa but its precise
position is unstable. In Bayesian analysis it is sister to holophyletic
Glycostylida, but distance trees mostly place it between Dermamoeba and
a possibly artifactual long-branch cluster including Thecamoeba. These
positions are poorly supported and basal amoebozoan branching illresolved, making it unclear whether Discosea (Glycostylida,
Himatismenida, Dermamoebida) is holophyletic; however, Thecamoeba
seems not specifically related to Dermamoeba. We also sequenced the
small-subunit rRNA gene of Vannella persistens, which constantly
grouped with other Vannella species, and two Hartmannella strains. Our
trees suggest that Vexilliferidae, Variosea and Hartmannella are
polyphyletic, confirming the existence of two very distinct
Hartmannella clades: that comprising H. cantabrigiensis and another
divergent species is sister to Glaeseria, whilst Hartmannella
vermiformis branches more deeply.
PMID: 16171188 [PubMed - in process]
32: Protist. 2005 Aug;156(2):191-202.
The testate lobose amoebae (order Arcellinida Kent, 1880) finally find
their home within Amoebozoa.
Nikolaev SI, Mitchell EA, Petrov NB, Berney C, Fahrni J, Pawlowski J.
Department of Evolutionary Biochemistry, A. N. Belozersky Institute of
Physico-Chemical Biology, Moscow State University, Moscow, Russia.
s-nikol@yandex.ru
Testate lobose amoebae (order Arcellinida Kent, 1880) are common in all
aquatic and terrestrial habitats, yet they are one of the last higher
taxa of unicellular eukaryotes that has not found its place in the tree
of life. The morphological approach did not allow to ascertain the
evolutionary origin of the group or to prove its monophyly. To solve
these challenging problems, we analyzed partial small-subunit ribosomal
RNA (SSU rRNA) genes of seven testate lobose amoebae from two out of
the three suborders and seven out of the 13 families belonging to the
Arcellinida. Our data support the monophyly of the order and clearly
establish its position among Amoebozoa, as a sister-group to the clade
comprising families Amoebidae and Hartmannellidae. Complete SSU rRNA
gene sequences from two species and a partial actin sequence from one
species confirm this position. Our phylogenetic analyses including
representatives of all sequenced lineages of lobose amoebae suggest
that a rigid test appeared only once during the evolution of the
Amoebozoa, and allow reinterpretation of some morphological characters
used in the systematics of Arcellinida.
PMID: 16171186 [PubMed - in process]
33: Nucleic Acids Res. 2005 Sep 16;33(16):5262-70. Print 2005.
Localization, mobility and fidelity of retrotransposed Group II introns
in rRNA genes.
Conlan LH, Stanger MJ, Ichiyanagi K, Belfort M.
Wadsworth Center, Center for Medical Science, New York State Department
of Health, 150 New Scotland Avenue, Albany, NY 12208, USA.
We previously showed that the group II Lactococcus lactis Ll.LtrB
intron could retrotranspose into ectopic locations on the genome of its
native host. Two integration events, which had been mapped to unique
sequences, were localized in the present study to separate copies of
the six L.lactis 23S rRNA genes, within operon B or D. Although further
movement within the bacterial chromosome was undetectable, the
retrotransposed introns were able to re-integrate into their
original homing site provided on a plasmid. This finding indicates not
only that retrotransposed group II introns retain mobility properties,
but also that movement occurs back into sequence that is heterologous
to the sequence of the chromosomal location. Sequence analysis of the
retrotransposed introns and the secondary mobility events back to the
homing site showed that the introns retain sequence integrity. These
results are illuminating, since the reverse transcriptase (RT) of the
intron-encoded protein, LtrA, has no known proofreading function, yet
the mobility events have a low error rate. Enzymatic digests were used
to monitor sequence changes from the wild-type intron. The results
indicate that retromobility events have approximately 10(-5)
misincorporations per nucleotide inserted. In contrast to the high RT
error rates for retroviruses that must escape host defenses, the
infrequent mutations of group II introns would ensure intron spread
through retention of sequences essential for mobility.
PMID: 16170154 [PubMed - indexed for MEDLINE]
34: J Biotechnol. 2005 Sep 13; [Epub ahead of print]
Performance and microbial communities of a continuous stirred tank
anaerobic reactor treating two-phases olive mill solid wastes at low
organic loading rates.
Rincon B, Raposo F, Borja R, Gonzalez JM, Portillo MC, Saiz-Jimenez C.
Instituto de la Grasa, CSIC, Avda. Padre Garcia Tejero 4, 41012
Sevilla, Spain.
A study of the performance and microbial communities of a continuous
stirred tank reactor (CSTR) treating two-phases olive mill solid wastes
(OMSW) was carried out at laboratory-scale. The reactor operated at a
mesophilic temperature (35 degrees C) and an influent substrate
concentration of 162g total chemical oxygen demand (COD)L(-1) and 126g
volatile solids (VS)L(-1). The data analyzed in this work corresponded
to a range of organic loading rates (OLR) of between 0.75 and 3.00g
CODL(-1)d(-1), getting removal efficiencies in the range of 97.0-95.6%.
Methane production rate increased from 0.164 to 0.659L
CH(4)L(reactor)(-1)d(-1) when the OLR increased within the tested
range. Methane yield coefficients were 0.225L CH(4)g(-1) COD removed
and 0.290L CH(4)g(-1) VS removed and were virtually independent of the
OLR applied. A molecular characterization of the microbial communities
involved in the process was also accomplished. Molecular identification
of microbial species was performed by PCR amplification of 16S
ribosomal RNA genes, denaturing gradient gel electrophoresis (DGGE),
cloning and sequencing. Among the predominant microorganisms in the
bioreactor, the Firmicutes (mainly represented by Clostridiales) were
the most abundant group, followed by the Chloroflexi and the GammaProteobacteria (Pseudomonas species as the major representative). Other
bacterial groups detected in the bioreactor were the Actinobacteria,
Bacteroidetes and Deferribacteres. Among the Archaea, the methanogen
Methanosaeta concilii was the most representative species.
PMID: 16168509 [PubMed - as supplied by publisher]
35: Parasitol Res. 2005 Sep 16; [Epub ahead of print]
Enterocytozoon bieneusi genotypes in dairy cattle in the eastern United
States.
Santin M, Trout JM, Fayer R.
Environmental Microbial Safety Laboratory, United States Department of
Agriculture, Animal and Natural Resources Institute Agricultural
Research Service, Building 173, BARC-East, 10300 Baltimore Avenue,
Beltsville, MD, 20705, USA, rfayer@anri.barc.usda.gov.
Fecal specimens were obtained from 12-24-month-old dairy heifers on
farms in Vermont, New York, Pennsylvania, Maryland, Virginia, North
Carolina, and Florida. PCR positive specimens for Enterocytozoon
bieneusi were found in 131 of 571 heifers examined (23%) and on all the
farms visited. The prevalence of E. bieneusi varied considerably across
farms, with the lowest prevalence (4.7%) on MD-2 and the highest
prevalence (37.8%) on NY-2. All PCR positive specimens that amplified
the ITS region as well as a portion of the flanking large and small
subunit ribosomal RNA genes were sequenced to determine the genotype(s)
of the E. bieneusi present and six genotypes were identified. Most were
identified as cattle-specific genotypes, previously reported from
cattle as BEB1, BEB2, BEB3, and BEB4. Two isolates were genetically
identical or similar to E. bienesusi reported as the human pathogens
Peru 6 and Peru 9 (or D) genotypes. Although our data demonstrate the
presence of zoonotic genotypes in cattle, most genotypes found in
cattle were host specific.
PMID: 16167161 [PubMed - as supplied by publisher]
45: Nucleic Acids Res. 2005 Sep 9;33(16):5106-11. Print 2005.
Constraining ribosomal RNA conformational space.
Favaretto P, Bhutkar A, Smith TF.
BioMolecular Engineering Research Center, Boston University, 36
Cummington Street, Boston, MA 02215, USA.
Despite the potential for many possible secondary-structure
conformations, the native sequence of ribosomal RNA (rRNA) is able to
find the correct and universally conserved core fold. This study
reports a computational analysis investigating two mechanisms that
appear to constrain rRNA secondary-structure conformational space:
ribosomal proteins and rRNA sequence composition. The analysis was
carried out by using rRNA-ribosomal protein interaction data for
the Escherichia coli 16S rRNA and free energy minimization software for
secondary-structure prediction. The results indicate that selection
pressures on rRNA sequence composition and ribosomal protein-rRNA
interaction play a key role in constraining the rRNA secondary
structure to a single stable form.
PMID: 16155182 [PubMed - indexed for MEDLINE]
51: Nucleic Acids Res. 2005 Sep 6;33(15):4995-5005. Print 2005.
An evolutionary intra-molecular shift in the preferred U3 snoRNA
binding site on
pre-ribosomal RNA.
Borovjagin AV, Gerbi SA.
Department of Molecular Biology, Cell Biology and Biochemistry, Brown
University, Division of Biology and Medicine, Providence, RI 02912,
USA.
Correct docking of U3 small nucleolar RNA (snoRNA) on pre-ribosomal RNA
(pre-rRNA) is essential for rRNA processing to produce 18S rRNA. In
this report, we have used Xenopus oocytes to characterize the
structural requirements of the U3 snoRNA 3'-hinge interaction with
region E1 of the external transcribed spacer (ETS) of pre-rRNA. This
interaction is crucial for docking to initiate rRNA processing. 18S
rRNA production was inhibited when fewer than 6 of the 8 bp of the U3
3'-hinge complex with the ETS could form; moreover, base pairing
involving the right side of the 3'-hinge was more important than the
left. Increasing the length of the U3 hinge-ETS interaction by 9 bp
impaired rRNA processing. Formation of 18S rRNA was also inhibited by
swapping the U3 5'- and 3'-hinge interactions with the ETS or by
shifting the base pairing of the U3 3'-hinge to the sequence directly
adjacent to ETS region E1. However, 18S rRNA production was partially
restored by a compensatory shift that allowed the sequence adjacent to
the U3 3'-hinge to pair with the eight bases directly adjacent to ETS
region E1. The results suggest that the geometry of the U3 snoRNA
interaction with the ETS is critical for rRNA processing.
PMID: 16147982 [PubMed - indexed for MEDLINE]