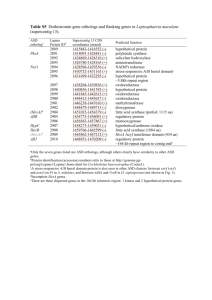
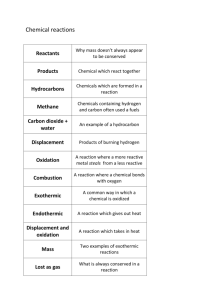
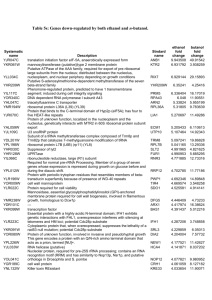
Biological Sciences/Evolution - Environmental Biophysics and
advertisement

Biological Sciences/Evolution Genome Evolution in Cyanobacteria: the Stable Core and the Variable Shell Tuo Shi*† and Paul G. Falkowski*‡§ *Environmental Biophysics and Molecular Ecology Program, Institute of Marine and Coastal Sciences, and ‡Department of Geological Sciences, Rutgers, The State University of New Jersey, New Brunswick, NJ 08901, USA 26 text pages, 7 figures and 3 tables 188 words in the abstract and total 36,149 characters in paper Author contribution: T.S. and P.G.F. designed research; T.S. performed research; T.S. and P.G.F. analyzed data; and T.S. and P.G.F. wrote the paper. The authors declare no conflict of interest. Abbreviations: LGT, lateral gene transfer; NJ, neighbor joining; ML, maximum likelihood; MP, maximum parsimony; PCoA, principal coordinates analysis; †Present §To address: University of California, Santa Cruz, 1156 High Street, Santa Cruz, CA 95064 whom correspondence should be addressed. Paul G. Falkowski, 71 Dudley Road, New Brunswick, NJ 08901. E-mail: falko@marine.rutgers.edu, Telephone: (732)-932-6555 ext.370, Fax: (732)-932-4083. Page 1 Abstract Cyanobacteria are the only known prokaryotes that perform oxygenic photosynthesis, the evolution of which transformed the biology and geochemistry of Earth. The rapid increase in published genomic sequences of cyanobacteria provides the first opportunity to reconstruct events in the evolution of oxygenic photosynthesis on the scale of entire genomes. Here we demonstrate the overall phylogenetic incongruence among 682 orthologous protein families from 13 genomes of cyanobacteria. However, principle coordinate analyses on the evolutionary relationships reveal a core set of 323 genes with similar evolutionary trajectories. The core set is extremely conservative in protein variability, and is made largely of genes encoding the major components in photosynthetic and ribosomal apparatus. Many of the key proteins are encoded by genome-wide conserved small gene clusters which are indicative of protein-protein, protein-prosthetic group and protein-lipid interactions. We propose that the macromolecular interactions in complex protein structures and metabolic pathways retard the tempo of evolution of the core genes, and hence exert a selection pressure that restricts piecemeal lateral gene transfer of components of the core. Identification of the core establishes a foundation for reconstructing robust organismal phylogeny in genome space. Keywords: cyanobacteria | gene family | lateral gene transfer | genome evolution | oxygenic photosynthesis Genome Evolution in Cyanobacteria Page 2 Introduction Cyanobacteria are the only oxygenic photosynthetic prokaryotes. Approximately 2.4 billion years ago, the evolution of that energy transduction pathway transformed Earth’s atmosphere and upper ocean, ultimately facilitating the development of complex life forms that are dependent on aerobic metabolism (1-3). Molecular fossils of cyanobacteria date back at least 3 Ga (4-8), and the clade has evolved into one of the largest and most diverse groups of bacteria on this planet today (9). Cyanobacteria contribute significantly to global primary production (10, 11), and diazotrophic taxa are central to global nitrogen cycle (12-14). Arguably, this group, more than any other prokaryotic phylum, has had a greater impact on the biogeochemistry and evolutionary trajectory on Earth, yet its own evolutionary history is poorly understood. The availability of complete sequences of genomes for clusters of related organisms provides the first opportunity to reconstruct events of genomic evolution through the analysis of entire functional classes (15). Currently, cyanobacteria represent one of the densest clusters of fully sequenced genomes (Table 1). Comparisons of genome sequences of closely related marine Prochlorococcus and Synechococcus species have demonstrated an intimate link between genome divergence in specific strains and their physiological adaptations to different oceanic niches (16, 17). This ecotypic flexibility appears to be driven by myriad selective pressures that govern genome size, GC content, gene gain and loss, and rate of evolution (18, 19). Moreover, phylogenetic analyses of genes shared by all the five known phyla of photosynthetic bacteria, including cyanobacteria (the only oxygenic group), purple bacteria (Proteobacteria), green sulfur bacteria (Chlorobi), green filamentous bacteria (Chloroflexi), and the gram-positive Genome Evolution in Cyanobacteria Page 3 heliobacteria (Firmicutes), have provided important insights into the origin and evolution of photosynthesis, an intensively debated subject in the past decades (20-31). This information has been substantially extended by genome-wide comparative informatics (32-34). One of the major implications of the latter work is a significant extent of lateral gene transfer (LGT) among these photosynthetic bacteria. The observation that cyanophages sometimes carry photosynthesis genes (35-38) provides one mechanism of rapid LGT among these phyla. However, LGTs almost certainly do not occur with equal probability for all genes. For example, informational genes (those involved in transcription, translation, and related processes), which are thought to have more macromolecular interactions than operational genes (those involved in housekeeping), are seldom transferred (39, 40). The existence of a core of genes resistant to LGT has been reported in recent studies using relatively intensive taxon sampling (41, 42), suggesting that there exists a set of genes that remain closely associated, resistant to LGT, and with an extremely slow evolutionary tempo. Identification of such core genes potentially allows separatation of true phylogenetic signals from “noise”. It is, therefore, of considerable interest to transcribe all coherent genome data into pertinent phylogenetic information (43) and to identify which genes are more susceptible to lateral transfer. Here we report on identification and reconstruction of the phylogeny of 682 orthologous protein families from 13 genomes of cyanobacteria. Our primary goals are twofold: a) to examine the impact of LGT on the evolution of photosynthesis and the radiation of cyanobacterial lineages; and b) to identify a core set of genes that are resistant to LGT on which robust organismal phylogy can be reconstructed. Our results reveal that >52% (359) of the orthologs are susceptible to LGT and hence are responsible Genome Evolution in Cyanobacteria Page 4 for the inconsistant phylogenetic signal of this taxon in genome space. In contrast, the remaining 323 othologs show broad phylogenetic agreement. This core set is comprised of key photosynthetic genes as well as those coding for ribosomal translational apparatus. This observation suggests that the macromolecular interactions in complex protein structures (e.g., ribosomal proteins) and metabolic pathways (e.g., oxygenic photosynthesis) are strongly resistant to piecemeal lateral gene transfer. Transfer was ultimately accomplished by wholesale incorporation of cyanobacteria into eukaryotic host cells, giving rise to primary photosynthetic endosymbionts which retained both photosynthetic genes and genes encoding for its own ribosome (44-46). Genome Evolution in Cyanobacteria Page 5 Results Conserved protein families in genomes of cyanobacteria Our pair-wise genome comparison revealed a total of 682 orthologous proteincoding genes that are common to all 13 genomes examined [supporting information (SI) Table 2]. These genes constitute the core gene set and some of these define aspects of the genotype that are uniquely cyanobacterial. This core set represents only 9.3% (in the case of the largest genome Nostoc punctiforme) to 39.8% (in the case of the smallest genome Prochlorococcus marinus MED4) of the total number of protein-coding genes from each genome under study (see Table 1), but seems to account for all of the principal functions in genome replication, expression, and repair, as well as the majority of the reactions in several central metabolic pathways (SI Table 3). Our analysis is based on identification of reciprocal best hits with a moderately customized BLAST e-value threshold (10e−4) compared to what has been used by others (18, 19). This leads to an estimate of the pool of orthologs similar to what has been identified from 10 cyanobacterial genomes (47), but nearly three times more than the number of cyanobacterial signature genes bioinformatically characterized by Martin et al. (48), and only 65% of the number of cyanobacterial clusters of orthologous groups (34). The discrepancy mostly results, in the case of the former, from a filtering procedure to remove homologs to chloroplasts and anoxygenic photosynthetic bacterial genomes, and in the case of the latter, from a less stringent, unidirectional BLAST hit scheme employed. In addition, some of the incomplete genomes used in this study are still undergoing confirmation from the final assembly, and it is possible that equivalent genes have been overlooked in some cases, resulting in an incomplete signature set. It is highly possible Genome Evolution in Cyanobacteria Page 6 that both reciprocal and unidirectional, circular best BLAST hit schemes result in an elimination of a large set of homolog families that are considered orthologs because of the overly restrictive criterion (49). However, the reciprocal best hit criterion detects more orthologs while producing the same qualitative results as the single best hit approach (47). Indeed, even without the use of any particular threshold (i.e., the default BLAST e-value threshold is 10), the set of orthologs identified via the reciprocal top BLAST hit scheme probably underestimates the actual number of orthologs (19). Phylogenetic incongruence among conserved protein families Based on amino acid sequences, we built phylogenetic trees for each of the 682 orthologous protein families using both neighbor joining (NJ) and maximum likelihood (ML) methods. To extrapolate major evolutionary trajectories, we analyzed and compared the observed topologies from all 682 individual trees. Surprisingly, the frequency distribution of observed topologies among the orthologous protein families failed to reveal a predominant, unanimous topology that represents a large number of orthologs (Fig. 1). In contrast, most of the orthologs (58% and 67% for NJ and ML, respectively) select a particular topology by itself. As a result, the maximum number of orthologs that share a particular topology is 15 and 13 for NJ and ML, respectively, which accounts for only 1.9% to 2.1% of the orthologous datasets (Fig. 1). This result clearly demonstrates the incongruence in phylogenetic information from individual protein families, even though they are highly conserved among cyanobacterial taxa. Phylogenomic reconcilement To determine whether genes give completely incompatible phylogenetic information or whether a common signal can be extracted from cyanobacterial Genome Evolution in Cyanobacteria Page 7 phylogenies, we employed a series of phylogenomic approaches, including the consensus of 682 individual trees, supertree reconstruction, as well as the reconstruction of phylogeny based on a concatenated alignment of 192,423 sites from the 682 sequences. These approaches greatly resolved the topological incongruence, leading to five topologies as shown in Fig. 2. Specifically, the NJ and ML trees built with different models using the concatenated sequences gave three topologies in total (T1 and T2 for NJ; T2 and T3 for ML), one of which is in agreement with that of the small subunit ribosomal RNA (SSU rRNA) tree repeatedly obtained with NJ, ML or maximum parsimony (MP) methods. The consensus- and super-tree built on the 682 individual NJ trees showed two other topologies (T4 and T5), whereas both the consensus- and supertree of ML trees revealed an identical topology to one of the concatenated ML trees. These five topologies are remarkably similar in that Synechocystis sp. PCC 6803 (SYN, see Table 1 for abbr. for other species) and five diazotrophic species form a monophyletic clade ((SYN,CWA),(TER,(NPU,(ANA,AVA)))), and that Synechococcus WH8102 (SYW) and three Prochlorococcus species form another monophyletic clade in three different formats: (SYW,(PMT,(PMS,PMM))), ((SYW,PMT),(PMS,PMM)), and (PMM,(PMS,(PMT,SYW))). The notable conflicts concern the species Synechococcus elongates PCC 7942 (SCO) and the thermophilic Thermosynechococcus elongates BP-1 (TEL), which tend to cluster at the base of the two major conserved subgroups but form aberrant topologies. However, analyses of the fitness of a particular topology to the 682 sequence alignments (SI Figs 6 and 7) indicated that almost all (97.5% to 99.6%) of the datasets support topologies T1-T5 at the 95% confidence level (P=0.95). This result suggests that Genome Evolution in Cyanobacteria Page 8 artificial paralogs or genes potentially obtained by LGT may thwart the establishment of organismal relationships based on plural consensus of individual gene phylogenies. The stable core and the variable shell in genome space To characterize the evolutionary trajectories with regarding to remove artificial paralogs, or genes potentially obtained by LGT, we calculated the tree distances among all possible pairs of the orthologous sets. The pair-wise distances were then used to conduct a principle coordinate analysis (PCoA). This resulted in a core set of 323 genes that share similar evolutionary histories (i.e., co-evolving genes) as opposed to the other 359 that exhibit divergent phylogenies (i.e., independently evolving genes) (Fig. 3). It is very striking that informational genes, especially those encoding ribosomal structural components, are almost all grouped in the densest region, while the tail is formed largely by operational genes. This observation is consistent with previous studies (39, 40) that indicate phylogenetic information is highly conserved in informational genes. Additionally, the core is comprised of key components constituting the scaffolds of the photosynthetic apparatus, and proteins that participate in chlorophyll biosynthesis and the Calvin cycle. Using the sum of all possible pairwise genetic distance as a measure of the protein variability (44), we compared the rates of evolution of genes in different functional categories. A plot of frequency distribution of genes versus protein variability (Fig. 4) reveals that ribosomal and photosynthetic genes are extremely conserved, whereas the operational and other informational genes were strongly skewed toward higher protein variability. This pattern of segregation is concordant with whether or not the genes are located in the core, and whether or not they are organized in conserved gene clusters. Genome Evolution in Cyanobacteria Page 9 This result suggests that the core gene set appears to have remained relatively stable throughout the entire evolutionary history of cyanobacteria, while genes in the shell are more likely to be acquired via LGT and are hence poorly conserved. Finally, we chose the 323 core genes to reconstruct the organismal phylogeny. The resulting tree, based on the concatenated core gene set, had the same topology as that obtained from the concatenation of all the 682 genes and that for the consensus and supertree of the trees obtained from all protein families (T3 in Fig. 2 and tree presented in Fig. 5). It differed only slightly from other tested topologies that are not rejected by many individual alignments (Fig. 2). Interestingly, the genomes from all the diazotrophic cyanobacteria cluster as one main group; this clade appears to have acquired its nitrogen fixation ability from a unicellular, thermophilic non-diazotrophic ancestor, probably during the Archaean Eon. The early diazotrophs almost certainly were non-heterocystous and eventually developed that differentiated cell type probably as a result of elevated level of atmospheric O2 (50). Genome Evolution in Cyanobacteria Page 10 Discussion Our analyses reveal an overwhelming phylogenetic discordance among the set of genes selected as likely orthologs (Fig. 1). Conflicting phylogenies can be a result of either artifacts of phylogenetic reconstruction, LGT or unrecognized paralogy. In our reciprocal best hit approach, we retained as orthologous gene families those containing only one gene per species. Therefore, only orthologous replacement and hidden paralogies (i.e., differential loss of the two copies in two lineages) can occur in selected families. These two types of events are expected to be comparatively rare under strict application of the reciprocal hit criterion (51). Thus, phylogenetic incongruence is unlikely due to artifacts resulting from a biased selection of orthologous genes. Furthermore, despite the overall disagreement in phylogenetic signal, most of the trees of individual proteins gave unambiguously strong support for at least some particular subtopologies which are repeatedly obtained through consensus, supertree and even concatenation analyses (Fig. 2). This finding obviates that phylogenetic incongruence is a result of tree reconstruction artifacts or long branch attraction effects. LGT is likely one of the most important driving forces that lead to the discrete evolutionary histories of the conserved protein families. Indeed, LGT has played an important role in the evolution of prokaryotic genomes (52-54). A hallmark of LGT is that the transferred genes often exhibit phylogenic incongruity and/or aberrant organismal distributions, which contrasts with the relationships inferred from both the SSU rRNA tree and other most resolved phylogenies of vertically inherited individual protein-coding genes. But how can this superficially random gene transfer event explain the conserved Genome Evolution in Cyanobacteria Page 11 nature of many of the key genes that comprise the functional core across all cyanobacterial taxa? While phylogenomic approaches are capable of capturing the consensus and the most frequently appeared bipartitions in individual phylogenies, leading to highly conserved subgroups or clusters that silhouette the major trend in genome evolution, they may not necessarily guarantee the paucity of a conflicting phylogenetic signal in genome space. The plural support for the consensus / supertree / concatenation topologies indicates that the five top topologies are not significantly different from each other at P=0.95; or in other words, all the five topologies would be accepted by more than 90% of the dataset using the same criterion because the data do not discriminate among the topologies. Do the consensus / supertree / concatenation trees accurately reflect organismal history? Or, on the contrary, do they blur the vertical inheritance signal by incorporating potential LGT? There is a large margin of uncertainty. Part of the uncertainty may be due to the strength of the SH test, especially when examining the accuracy of similar topologies. Indeed, the SH test is based on the evaluation of only 15 candidate topologies out of a total of 13,749,310,575 possible unrooted tree topologies for 13 species. Although the majority of the 13,749,310,575 possible topologies would not be supported by any dataset, the selection of a limited number of trees may have biased the analyses. But part of the uncertainty can also be attributed to the data, most notably the poorly conserved proteins that are disturbed by LGT. This is even more pronounced in the PCoA, which demonstrates clearly that about 53% of the orthologs are subject to frequent lateral transfers that might have complicated/diluted the vertical inheritance signal (Fig. 3). Genome Evolution in Cyanobacteria Page 12 Our results reveal a core set of genes in cyanobacteria that share similar evolutionary histories. The pattern of co-evolution of these genes is concomitant with extraordinarily conserved protein primary structure (Figs. 4 and 5). This finding of limited lateral transfer in highly conserved genes is consistent with the complexity hypothesis; that is, genes coding for large complex systems that have more macromolecular interactions are less subject to horizontal transfer than genes coding for small assemblies of a few gene products (40). In addition to the ribosomal genes, a significant number of photosynthetic genes are also clustered in the core, suggesting that photosynthetic genes may be as constrained as the ribosomal genes through evolution. To function effectively, the cyanobacterial photosynthetic apparatus needs an investment of a huge number of proteins, pigments, cofactors, and trace elements. Moreover, genes coding for the photosynthetic apparatus are scattered along the chromosome as small clusters of two to four genes that are highly conserved among all cyanobacteria (55). Although large-scale gene order has been shown to be relatively unstable in the prokaryotic genomes (56-58), the conservation of a minimum number of essential operons encoding proteins that function together in a pathway or structural complex appears to prevent genes from being perturbed by LGT (55, 59). The complexity of oxygenic photosynthetic machinery makes it difficult to transfer components piecemeal to non-photosynsthetic prokaryotes. Indeed, operon splitting of the photosynthetic apparatus requires many independent transfers of noncontiguous operons. Although large-scale horizontal gene transfer among photosynthetic prokaryotes (32) may suggest a complex non-linear process of evolution that results in a mosaic structure of photosynthetic pathway (60), transfers of the key photosynthetic genes are very rare (36). Genome Evolution in Cyanobacteria Page 13 Transfer of this key pathway was only achieved by wholesale incorporation of cyanobacteria into eukaryotic host cells (44-46). Genome Evolution in Cyanobacteria Page 14 Materials and Methods Gene Family Selection We performed all-against-all BLAST (61) comparisons of protein sequences for all possible pairs of the 13 genomes of cyanobacteria (Table 1) using an e value of 10−4 as a lower limit cutoff, and reciprocal genome-specific best hits were identified. We considered genes as being probable orthologs when they were included in an assembled set comprising the 13 genomes in which each gene was each other’s best BLAST match. A total number of 682 protein families consisting of one gene per genome were retrieved and assigned to functional categories according to those defined for the cluster of orthologous groups (COGs) (62). Because of the lack of a particular category for photosynthetic genes, the latter were assigned to the “energy production and conversion” COG category. Other genes which fell into more than one of the 19 COG categories have been assigned to a supplemental category called “miscellaneous” (19). Alignments and Tree Construction Protein sequences were aligned with ClustalW (63) with manual corrections, followed by selecting unambiguous parts of the alignments and concatenating sequences excluding all gap sites with Gblocks program (64). The maximum likelihood (ML) trees were computed for each family with the PHYML program (65) using the Jones, Taylor, and Thornton (JTT) model of substitution (66) and the γ-based method for correcting the rate heterogeneity among sites. For each family, the neighbor joining (NJ) trees were also constructed using the distance matrix provided by TREE-PUZZLE program version 5.1 (67) under a γ-based model of substitution (alpha parameter estimated by PUZZLE, eight Genome Evolution in Cyanobacteria Page 15 γ rate categories) and bootstrapped using SEQBOOT and CONSENSE from the PHYLIP package version 3.6 (68). Concatenation, consensus and supertree To generate a set of reasonable candidate topologies that could be tested against the alignments for individual gene, we used the consensus of the 682 trees from individual protein families (two methods, yielding topologies T3 and T4 in Fig. 2), the concatenation of all the proteins (192,423 amino acids) (seven methods, yielding topologies T1 to T3), the supertree (two methods, yielding topologies T3 and T5), and the SSU rRNA (three methods, yielding topology T1). In the case of the reconstruction of the trees based on the SSU rRNA, we used the PHYLO_WIN (69) to perform the maximum parsimony (MP), NJ with bootstrap, ML reconstruction using the γ-based method for correcting the rate heterogeneity among sites. The consensus of the trees of the 682 protein alignments was obtained using the CONSENSE module of the PHYLIP package. As there are no missing data, we also concatenated all the proteins and constructed the tree either using the PROTML program in the MOLPHY package (70) or using the quartet puzzling (QP) implemented with TREE-PUZZLE. Trees chosen for the supertree computation were coded into a binary matrix using the coding scheme of Baum (71) and Ragan (72). The matrices obtained were concatenated into a supermatrix. The supertree was calculated on the supermatrix using program DNAPARS (default options) from the PHYLIP package. Bootstrap values on the supermatrix were obtained using SEQBOOT and CONSENSE. Comparisons between Trees Genome Evolution in Cyanobacteria Page 16 Trees were compared with Treedist program using the branch score distance of Kuhner and Felsenstein (73). On the nn distance matrix obtained (n is the number of trees), a principle coordinates analysis (PCoA) is computed using SAS software. PCoA, a multivariate ordination method based on distance matrices, allowed us to embed our n trees in a space of up to n-1 dimensions. By taking the most significant two first dimensions and plotting the objects (the trees) along these, the major trends and groupings in the data can be determined by visual inspection. For each of the 682 alignments, a comparison of the likelihood of the best topology with that of the candidate topologies (SI Fig. 6) was performed with the Shimodaira-Hasegawa (SH) test (74) implemented in TREE-PUZZLE. This test determines whether these potential organismal phylogenies are significantly rejected by the alignment and thus whether LGT or some other anomalous process must be invoked. Genome Evolution in Cyanobacteria Page 17 Acknowledgements We thank William Martin and Colomban de Vargas for valuable comments and stimulating discussions and Lin Jiang for help with the principle coordinates analysis. This research is supported by NASA grants to P.G.F.. Sequence alignments and tree files are available upon request. Genome Evolution in Cyanobacteria Page 18 References 1. Blankenship, R. E. & Hartman, H. (1998) Trends in Biochemical Sciences 23, 94-97. 2. Falkowski, P. G., Katz, M. E., Milligan, A. J., Fennel, K., Cramer, B. S., Aubry, M. P., Berner, R. A., Novacek, M. J. & Zapol, W. M. (2005) Science 309, 2202-2204. 3. Raymond, J. & Segre, D. (2006) Science 311, 1764-1767. 4. Schopf, J. & Packer, B. (1987) Science 237, 70-73. 5. Buick, R. (1992) Science 255, 74-77. 6. Brocks, J. J., Logan, G. A., Buick, R. & Summons, R. E. (1999) Science 285, 10331036. 7. Bekker, A., Holland, H. D., Wang, P. L., Rumble, D., Stein, H. J., Hannah, J. L., Coetzee, L. L. & Beukes, N. J. (2004) Nature 427, 117-120. 8. Knoll, A. H., Summons, R. E., Waldbauer, J. R. & Zumberge, J. E. (2007) in Evolution of primary producers in the sea, eds. Falkowski, P. G. & Knoll, A. H. (Academic Press, New York), pp. 134-163. 9. Whitton, B. A. & Potts, M. (2000) in The Ecology of Cyanobacteria, eds. Whitton, B. A. & Potts, M. (Kluwer Academic Publishers, Dordrecht, The Netherlands), pp. 1-11. 10. Waterbury, J. B., Watson, S. W., Guillard, R. R. L. & Brand, L. E. (1979) Nature 277, 293-294. 11. Chisholm, S. W., Olson, R. J., Zettler, E. R., Goericke, R., Waterbury, J. B. & Welschmeyer, N. A. (1988) Nature 334, 340-343. 12. Capone, D. G., Zehr, J. P., Paerl, H., Bergman, B. & Carpenter, E. J. (1997) Science 276, 1221-1229. Genome Evolution in Cyanobacteria Page 19 13. Zehr, J. P., Waterbury, J. B., Turner, P. J., Montoya, J. P., Omoregie, E., Steward, G. F., Hansen, A. & Karl, D. M. (2001) Nature 412, 635-638. 14. Karl, D., Michaels, A., Bergman, B., Capone, D., Carpenter, E., Letelier, R., Lipschultz, F., Paerl, H., Sigman, D. & Stal, L. (2002) Biogeochemistry 57/58, 47-98. 15. Koonin, E. V., Aravind, L. & Kondrashov, A. S. (2000) Cell 101, 573-576. 16. Palenik, B., Brahamsha, B., Larimer, F. W., Land, M., Hauser, L., Chain, P., Lamerdin, J., Regala, W., Allen, E. E., McCarren, J., Paulsen, I., Dufresne, A., Partensky, F., Webb, E. A. & Waterbury, J. (2003) Nature 424, 1037-1042. 17. Rocap, G., Larimer, F. W., Lamerdin, J., Malfatti, S., Chain, P., Ahlgren, N. A., Arellano, A., Coleman, M., Hauser, L., Hess, W. R., Johnson, Z. I., Land, M., Lindell, D., Post, A. F., Regala, W., Shah, M., Shaw, S. L., Steglich, C., Sullivan, M. B., Ting, C. S., Tolonen, A., Webb, E. A., Zinser, E. R. & Chisholm, S. W. (2003) Nature 424, 1042-1047. 18. Hess, W. R. (2004) Current Opinion in Biotechnology 15, 191-198. 19. Dufresne, A., Garczarek, L. & Partensky, F. (2005) Genome Biology 6, R14. 20. Olson, J. M. & Pierson, B. K. (1987) Orig Life 17, 419 - 430. 21. Blankenship, R. E. (1992) Photosynth Res 33, 91 - 111. 22. Vermaas, W. F. J. (1994) Photosynth Res 41, 285-294. 23. Xiong, J., Inoue, K. & Bauer, C. E. (1998) Proc Natl Acad Sci USA 95, 14851 14856. 24. Gupta, R. S., Mukhtar, T. & Singh, B. (1999) Mol Microbiol 32, 893 - 906. 25. Xiong, J., Fischer, W. M., Inoue, K., Nakahara, M. & Bauer, C. E. (2000) Science 289, 1724-1730. Genome Evolution in Cyanobacteria Page 20 26. Baymann, F., Brugna, M., Muhlenhoff, U. & Nitschke, W. (2001) Biochimica et Biophysica Acta (BBA) - Bioenergetics 1507, 291-310. 27. Gupta, R. S. (2003) Photosynth Res 76, 173-183. 28. Rutherford, A. W. & Faller, P. (2003) Phil. Trans R. Soc Lond. B. 358, 245-253. 29. Olson, J. M. & Blankenship, R. E. (2004) Photosynth Res 80, 373 - 386. 30. Sadekar, S., Raymond, J. & Blankenship, R. E. (2006) Mol Biol Evol 23, 2001 - 2007. 31. Xiong, J. (2006) Genome Biology 7, 245. 32. Raymond, J., Zhaxybayeva, O., Gogarten, J. P., Gerdes, S. Y. & Blankenship, R. E. (2002) Science 298, 1616-1620. 33. Zhaxybayeva, O., Hamel, L., Raymond, J. & Gogarten, J. (2004) Genome Biology 5, R20. 34. Mulkidjanian, A. Y., Koonin, E. V., Makarova, K. S., Mekhedov, S. L., Sorokin, A., Wolf, Y. I., Dufresne, A., Partensky, F., Burd, H., Kaznadzey, D., Haselkorn, R. & Galperin, M. Y. (2006) PNAS 103, 13126-13131. 35. Bailey, S., Clokie, M. R. J., Millard, A. & Mann, N. H. (2004) Research in Microbiology 155, 720-725. 36. Lindell, D., Sullivan, M. B., Johnson, Z. I., Tolonen, A. C., Rohwer, F. & Chisholm, S. W. (2004) PNAS 101, 11013-11018. 37. Millard, A., Clokie, M. R. J., Shub, D. A. & Mann, N. H. (2004) PNAS 101, 1100711012. 38. Sullivan, M. B., Lindell, D., Lee, J. A., Thompson, L. R., Bielawski, J. P. & Chisholm, S. W. (2006) PLoS Biology 4, e234. 39. Rivera, M. C., Jain, R., Moore, J. E. & Lake, J. A. (1998) PNAS 95, 6239-6244. Genome Evolution in Cyanobacteria Page 21 40. Jain, R., Rivera, M. C. & Lake, J. A. (1999) PNAS 96, 3801-3806. 41. Daubin, V., Gouy, M. & Perriere, G. (2002) Genome Res. 12, 1080-1090. 42. Lerat, E., Daubin, V. & Moran, N. A. (2003) PLoS Biology 1, 101-109. 43. Eisen, J. A. & Fraser, C. M. (2003) Science 300, 1706-1707. 44. Rujan, T. & Martin, W. (2001) Trends in Genetics 17, 113-120. 45. Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., Leister, D., Stoebe, B., Hasegawa, M. & Penny, D. (2002) Proc. Natl. Acad. Sci. USA 99, 1224612251. 46. Falkowski, P. G. & Knoll, A. H. (2007) Evolution of primary producers in the sea (Academic Press, New York). 47. Zhaxybayeva, O., Lapierre, P. & Gogarten, J. P. (2004) Trends in Genetics 20, 254260. 48. Martin, K., Siefert, J., Yerrapragada, S., Lu, Y., McNeill, T., Moreno, P., Weinstock, G., Widger, W. & Fox, G. (2003) Photosynthesis Research 75, 211-221. 49. Tatusov, R. L., Koonin, E. V. & Lipman, D. J. (1997) Science 278, 631-637. 50. Tomitani, A., Knoll, A. H., Cavanaugh, C. M. & Ohno, T. (2006) Prot. Natl. Acad. Sci. USA 103, 5442-5447. 51. Zhaxybayeva, O. & Gogarten, J. P. (2003) BMC Genomics 4, 37. 52. Ochman, H., Lawrence, J. G. & Groisman, E. A. (2000) Nature 405, 299-304. 53. Gogarten, J. P., Doolittle, W. F. & Lawrence, J. G. (2002) Mol Biol Evol 19, 22262238. 54. Boucher, Y., Douady, C. J., Papke, R. T., Walsh, D. A., Boudreau, M. E. R., Nesbo, C. L., Case, R. J. & Doolittle, W. F. (2003) Annual Review of Genetics 37, 283-328. Genome Evolution in Cyanobacteria Page 22 55. Shi, T., Bibby, T. S., Jiang, L., Irwin, A. J. & Falkowski, P. G. (2005) Mol Biol Evol 22, 2179-2189. 56. Mushegian, A. R. & Koonin, E. V. (1996) Trends Genet 12, 289–290. 57. Siefert, J. L., Martin, K. A., Abdi, F., Widger, W. R. & Fox, G. E. (1997) J Mol Evol 45, 467–472. 58. Itoh, T., Takemoto, K., Mori, H. & Gojobori, T. (1999) Mol Biol Evol 16, 332–346. 59. Dandekar, T., Snel, B., Huynen, M. & Bork, P. (1998) Trends in Biochemical Sciences 23, 324-328. 60. Blankenship, R. E. (2001) Trends in Plant Science 6, 4-6. 61. Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucl. Acids. Res. 25, 3389-3402. 62. Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., Kiryutin, B., Galperin, M. Y., Fedorova, N. D. & Koonin, E. V. (2001) Nucl. Acids Res. 29, 22-28. 63. Thompson, J., Higgins, D. & Gibson, T. (1994) Nucleic Acids Res. 22, 4673-4680. 64. Castresana, J. (2000) Mol. Biol. Evol. 17, 540-552. 65. Guindon, S. & Gascuel, O. (2003) Systematic Biology 52, 696-704. 66. Jones, D. T., Taylor, W. R. & Thornton, J. M. (1992) Computer Applications in the Biosciences (CABIOS) 8, 275-282. 67. Strimmer, K. & von Haeseler, A. (1996) Mol Biol Evol 13, 964-969. 68. Felsenstein, J. (2002). 69. Galtier, N., Gouy, M. & Gautier, C. (1996) Comput. Applic. Biosci. 12, 543-548. 70. Adachi, J. & Hasegawa, M. (1996) Comput. Sci. Monogr. No. 28, 1-150. Genome Evolution in Cyanobacteria Page 23 71. Baum, B. R. (1992) Taxon 41, 3–10. 72. Ragan, M. A. (1992) Mol Phylogenet Evol 1, 53-58. 73. Kuhner, M. K. & Felsenstein, J. (1994) Mol Biol Evol 11, 459-468. 74. Shimodaira, H. & Hasegawa, M. (1999) Mol Biol Evol 16, 1114-1116. Genome Evolution in Cyanobacteria Page 24 Tables Table 1 General features of the genomes of the 13 cyanobacteria used in this study Genome Abbr. Size (mb) (G+C) % Protein coding genes Database Reference Anabaena (Nostoc) sp. PCC7120 Anabaena variabilis ATCC29413 Crocosphaera watsonii WH8501 Gloeobacter violaceus PCC7421 Nostoc punctiforme ATCC29133 Prochlorococcus marinus MED4 Prochlorococcus marinus MIT9313 Prochlorococcus marinus SS120 Synechococcus elongatus PCC7942 Synechococcus sp. WH8102 Synechocystis sp. PCC6803 (Thermo) Synechococcus elongatus BP-1 Trichodesmium erythraeum IMS101 ANA AVA CWA GVI NPU PMM PMT PMS SCO SYW SYN TEL TER 6.41 7.10 6.30 4.66 9.50 1.66 2.41 1.75 2.70 2.43 3.57 2.59 6.50 42.5 41.4 37.1 64.0 41.4 32.0 50.8 36.0 55.0 59.5 47.7 54.0 34.2 5,366 5,754 6,751 4,430 7,281 1,713 2,267 1,884 2,876 2,526 3,264 2,475 5,266 CyanoBase JGI/DoE JGI/DoE CyanoBase JGI/DoE JGI/DoE JGI/DoE Roscoff JGI/DoE JGI/DoE CyanoBase CyanoBase JGI/DoE Kaneko et al., 01 Nakamura et al., 03 Meeks, 01 Rocap et al., 03 Rocap et al., 03 Dufresne et al., 03 Palenik et al., 03 Kaneko et al., 96 Nakamura et al., 02 Database website: CyanoBase (http://www.kazusa.or.jp/cyanobase/); JGI/DOE (http://www.jgi.doe.gov/); Roscoff (http://www.sb-roscoff.fr/) Genome Evolution in Cyanobacteria Page 25 Figure legends Fig.1. The distribution of tree topologies among 682 sets of orthologs. Both neighbor joining (NJ, black bars) and maximum likelihood (ML, red bars) tree topologies gave similar distribution pattern. There is no unanimous support for a single topology; rather most of the orthologs (58% and 67% for NJ and ML trees, respectively) appeared as singletons which associate with unique topologies. Fig. 2. Representative backbone tree topologies. Phylogenetic trees were constructed using both SSU rRNA gene and orthologous proteins through phylogenomic approaches (see Materials and Methods for details). The phylogenetic tree construction methods are highlighted with colored horizontal bars and text. The conserved monophyletic subgroups in each topology (T1-T5) are shaded. The top panel shows the proportion of orthologs giving a particular tree topology (NJ, black bar; ML, red bar). Also shown are the examples of proteins corresponding to that topology. The bottom panel indicates number of datasets accepting (left) or rejecting (right) a particular topology in a ShimodairaHasegawa (SH) (74) test of each of the backbone topologies along with other candidates randomly obtained (SI Fig. 7). Note that the consensus topology (T3) is rejected by only a few gene families. Species abbreviations as in Table 1. Fig. 3. Principal coordinates analysis (PCoA) of trees compared with topological distance. (A) Plot of the two first axes of the PCoA made from 628 ML trees. The other 54 genes are excluded as a result of axis demarcation. The same experiment with NJ trees gave very similar results. The ellipse depicts 323 orthologs in the densest region (the Genome Evolution in Cyanobacteria Page 26 core) of the cloud that share a common phylogenetic signal, while trees present in the marginal area (the shell) are much more likely to be perturbed by lateral transfers. Photosynthetic genes are color coded based on their respective pathways. Also shown are examples of conserved small clusters of ribosomal (red text) and photosynthetic (green text) genes that are located in the core. (B) The PCoA plotted against the protein variability. Photosynthetic genes are collectively designated as green dots. Note both the ribosomal and key photosynthetic genes are extremely conservative in protein variability. Fig. 4. Frequency distribution of the fraction of genes belonging to designated categories within each 1.0 interval of protein variability. Protein variability was measured by taking the total length of the corresponding tree obtained in the phylogenetic analysis as measured by amino acid substitutions, divided by the number of sequences in the tree. Fig. 5. The ML tree reconstructed based on the concatenation of the core 323 proteins. The topology shown agrees with the consensus topology of the 682 orthologs (T3 of Fig. 2) and is supported by almost all individual datasets (Fig. 2 and SI Fig. 7).The divergence of diazotrophic cyanobacteria is designated by the dashed line. Genome Evolution in Cyanobacteria Page 27 Figures Number of topologies 500 NJ ML 400 30 20 10 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 Number of orthologs Shi & Falkowski, Fig. 1 Genome Evolution in Cyanobacteria Page 28 Shi & Falkowski, Fig. 2 Genome Evolution in Cyanobacteria Page 29 1.0 A Operational Informational Ribosomal ATPase Calvin cycle Heme & Chlorophyll PSI PSII Cyt. b6f Dimension 2 0.5 NADH dehydrogenase Ycf 0.0 rpl20/35; rpl27/21; rpl3/4/23; rpl2/rps19; rpl33/rps18; rpl12/10; rpl22/rps3/rpl16; rps17/rplN/24/5/rps8; rpl6/18/rpsE/rpl15; rps13/11/rpoA/rpl17; rps12/7/fusA/tufA/rps10; -0.5 psbEF; petBD; psaAB; atpIH; atpAC; atpBE; ndhFD; ndhAIGE -1.0 -1.0 -0.5 0.0 0.5 1.0 Dimension 1 B High 12 Operational Informational Ribosomal 10 Low y Protein variabilit Photosynthetic 8 6 4 2 0 0.5 me Di 0.0 2 ion ns 1.0 -0.5 0.5 0.0 -1.0 -0.5 -1.0 sion 1 Dimen Shi & Falkowski, Fig. 3 Genome Evolution in Cyanobacteria Page 30 0.50 operational informational ribosomal photosynthetic Proportion 0.40 0.30 0.20 0.10 0.00 0.25 core shell Proportion 0.20 0.15 0.10 0.05 0.00 0.25 clustered unclustered Proportion 0.20 0.15 0.10 0.05 0.00 0 Low 2 4 6 8 10 Protein variability 12 High Shi & Falkowski, Fig. 4 Genome Evolution in Cyanobacteria Page 31 Shi & Falkowski, Fig. 5 Genome Evolution in Cyanobacteria Table 2 legend Page 1 Online Supporting Information Table 2. Ortholog table including 682 conserved protein families from 13 genomes of cyanobacteria. Column headings indicate ortholog ID, gene name, encoded protein, and cyanobacterial species used in this study. Species abbreviation: ANA, Anabaena sp. PCC 7120; AVA, Anabaena variabilis ATCC29413; CWA, Crocosphaera watsonii WH8501; GVI, Gloeobacter violaceus PCC 7421; NPU, Nostoc punctiforme; PMM, Prochlorococcus marinus MED4; PMS, Prochlorococcus marinus SS120; PMT, Prochlorococcus marinus MIT9313; SCO, Synechococcus elongatus PCC7942; SYW, Synechococcus sp. WH8102; SYN, Synechocystis sp. PCC 6803; TEL, Thermosynechococcus elongatus BP-1; TER, Trichodesmium erythraeum IMS101. For species whose genomes were in draft form when this work was undertaken, preliminary protein coding sequences were based on contig assemblies as of their respective release dates: NPU, April 9, 2001; SCO, September 22, 2003; TER, December 16, 2003; AVA, January 19, 2005; CWA, January 30, 2004. The letters and numbers in each column refer to the gene designators used by the individual genome projects, which are the likely ortholog detected by the analysis reported here. In some cases, e.g., Nostoc, the name consists of a contig followed by the gene number on that contig. In some cases the designator, e.g., sll0698, indicates a relative direction of transcription too. Genes are tabulated for each functional category in the physical order in which they occur in Synechococcus sp. WH8102. This allows the identification of genome-wide conserved gene clusters (putative operons) containing two or more signature genes by simply scanning the table. Signature genes included in these putative operons are tabulated in italic. Informational (purple), ribosomal (pink) and photosynthetic genes (green) are color shaded. Protein variability derived from the phylogenetic tree for each gene was Online Supporting Information Table 2 legend Page 2 indicated. The binary number designates whether protein families were found in the core (1, also in bold) or in the shell (0) of the genome as in Fig. 3. Online Supporting Information Table 2 Pag ID Prot. Var. Core / shell ort343 ort539 ort537 ort186 ort190 ort210 ort364 ort087 ort017 ort392 ort341 ort145 ort149 ort452 ort453 ort369 ort648 ort347 ort002 ort484 ort046 ort394 ort647 ort156 ort585 ort596 ort267 ort344 ort308 ort224 ort026 ort331 ort170 ort437 ort174 ort232 ort177 ort673 ort460 ort565 ort630 4.8597 3.5725 7.7917 8.7381 5.9564 8.0168 3.5384 5.0449 3.9641 4.3684 3.4126 6.9095 5.2881 7.0501 8.0446 2.7995 6.5608 2.2050 4.0014 5.6054 6.6183 2.0643 2.9453 7.2478 2.5480 4.1919 8.0685 5.2289 2.1164 7.6150 4.3987 5.6049 3.5064 5.2913 8.5078 10.3689 4.3443 2.7635 4.4651 2.7893 2.7558 1 1 0 0 0 0 1 0 1 0 1 0 1 0 0 1 0 1 1 0 0 1 1 0 1 1 0 0 1 0 1 1 1 1 0 0 0 1 0 1 1 ANA GVI PMM PMT NPU SCO PMS SYN TEL TER SYW AVA CWA Gene Product alr2010 all3652 all3651 all0443 alr4943 all1760 all1759 all1758 alr3887 alr2445 alr2447 alr2449 alr5067 alr5066 alr5065 all5062 alr5061 all5058 all5057 alr4755 all2847 all2846 all3519 alr3430 alr3428 asr3427 all4933 alr4932 alr3684 alr3681 all3680 all3679 all3678 alr3644 alr3444 all0932 all3013 all1132 alr3638 alr3993 alr4851 glr3232 glr2113 glr2114 gll1546 gll1464 gll1336 glr0251 glr1672 gll1051 gll4194 glr4267 glr4268 glr3498 glr2317 glr2316 glr0530 glr3298 glr1285 glr1286 glr0970 gll1629 glr2615 gll1807 gll0286 gll2784 gsl2785 glr2559 glr1609 glr4196 glr4195 glr1017 glr1018 glr1019 gll1774 gll0066 gll0401 glr4006 gll1855 gll1033 glr1042 gll1836 0001 0003 0004 0006 0007 0009 0010 0011 0012 0016 0017 0019 0020 0021 0022 0023 0024 0026 0027 0028 0036 0037 1664 1663 1662 1661 1659 1658 1657 1655 1654 1653 1652 1648 1647 1651 1650 1649 1645 1643 1639 0001 0003 0004 0006 0007 0010 0011 0012 0013 0021 0022 0024 0025 0026 0027 0028 0029 0031 0032 0033 0044 0045 0052 0054 0055 0056 0058 0059 0061 0063 0064 0065 0066 0073 0074 0068 0069 0071 0079 0081 0083 424.5 430.35 430.36 483.77 485.23 506.137 506.138 506.142 432.30 506.170 506.172 506.174 418.36 418.34 418.33 418.18 418.32 418.21 418.22 505.73 472.81 472.82 505.205 498.153 498.151 498.150 505.147 505.44 448.70 448.68 448.8 448.9 448.10 482.108 505.39 501.71 439.16 418.3 504.24 509.108 427.41 136.2572 136.2574 136.2575 136.2839 136.2838 129.527 129.528 129.529 129.531 135.1882 135.1884 135.1886 136.2277 135.2117 135.2116 135.1726 129.567 129.576 129.577 132.998 136.2428 136.2429 136.2864 128.510 128.507 128.506 134.1557 126.225 129.548 129.546 135.2061 135.2062 135.2063 134.1469 136.2783 134.1586 134.1395 136.2222 130.690 130.693 136.2714 0001 0003 0004 0006 0007 0009 0010 0011 0012 0016 0017 0019 0020 0021 0022 0023 0024 0026 0027 0028 0037 0038 1824 1823 1822 1821 1819 1818 1817 1815 1814 1813 1812 1808 1807 1811 1810 1809 1805 1803 1801 slr0965 sll1056 sll0757 sll0837 sll1348 sll0271 slr2102 slr2031 slr1133 sll0057 sll0897 sll0898 slr1847 slr1424 slr1423 sll1342 slr1787 slr0434 slr0435 sll0660 slr1538 slr0213 slr0633 slr0171 sll0767 ssl1426 sll1377 sll1360 sll0535 sll0533 slr0549 slr0550 slr0551 slr0657 sll0544 slr1278 slr2032 sll0459 sll1165 sll1282 sll0616 tll2349 tlr1683 tlr1684 tll2091 tlr0587 tll0787 tlr0928 tlr2236 tll0366 tlr1314 tll0790 tll0987 tlr0723 tll0370 tll0371 tll1466 tll2366 tlr1294 tlr1295 tll2045 tlr0767 tll2418 tll0065 tll2406 tll2158 tsl2159 tlr1854 tlr1132 tlr0510 tlr0508 tlr0069 tlr1219 tlr1220 tlr1833 tlr1573 tlr1616 tlr1249 tlr2033 tlr2240 tlr0067 tll1851 97.8656 122.6536 119.4070 118.3826 120.5163 118.3710 118.3711 118.3712 118.3713 111.1619 111.1622 111.1625 120.4678 120.4677 120.4676 120.4961 120.4675 112.1685 112.1688 95.8488 114.2291 114.2292 122.6785 119.4203 119.4201 119.4200 121.5205 116.2672 118.3957 118.3955 118.3578 118.3579 118.3580 96.8564 107.800 121.5631 122.7130 122.7522 119.4444 103.289 122.6969 0001 0003 0004 0006 0007 0010 0011 0012 0013 0023 0024 0026 0027 0028 0029 0030 0031 0033 0034 0035 0046 0047 0054 0056 0057 0058 0060 0061 0063 0065 0066 0067 0068 0070 0073 0075 0076 0077 0078 0082 0086 232.3907 232.2534 232.2535 232.1788 232.1191 232.4168 232.4169 232.4170 232.786 232.4265 232.4267 232.4269 232.1283 232.1282 232.1281 232.1279 232.1278 232.1276 232.1275 232.893 232.4920 232.4921 232.2129 232.2353 232.2351 232.2350 232.1179 232.1178 232.2502 232.2504 232.2505 232.2506 232.2507 232.2540 232.2371 232.4394 232.4766 232.3538 232.2097 232.686 232.1095 358.4593 358.4475 358.4479 361.5946 362.6447 359.4742 235.1403 241.1494 241.1495 278.2040 231.1351 361.5601 295.2370 361.5931 361.5930 201.980 211.1112 359.4728 359.4729 332.3322 362.6013 361.5637 362.6674 329.3240 361.5828 361.5827 358.4612 321.3017 319.2952 122.331 336.3450 336.3451 336.3452 284.2172 317.2914 267.1864 361.5613 361.5490 341.3608 346.3807 270.1905 dnaN purL purF DNA polymerase III beta subunit phosphoribosylformylglycinamidine synthetase II Glutamine amidotransferase class-II:Phosphoribosyl conserved hypothetical protein conserved hypothetical protein NusB family protein signal recognition particle docking protein FtsY conserved hypothetical protein Fumarate lyase:Delta crystallin putative heat shock protein GrpE DnaJ protein conserved hypothetical protein conserved hypothetical protein UDP-N-acetylenolpyruvoylglucosamine reductase Probable UDP-N-acetylmuramate-alanine ligase glyceraldehyde 3-phosphate dehydrogenase (NADP putative thiamine monophosphate kinase Translation elongation factor EF-P biotin carboxyl carrier protein (BCCP) subunit of ac putative pyridoxal phosphate biosynthetic protein Pdx cobalamin biosynthesis protein D GMP synthase (glutamine-hydrolysing) thiamin biosynthesis protein conserved hypothetical protein 50S ribosomal protein L20 50S ribosomal protein L35 putative glycosyltransferase DNA polymerase, gamma and tau subunits Putative ATP-dependent protease ATP-binding su Possible FKBP-type peptidyl-prolyl cis-trans isomera aspartate-semialdehyde dehydrogenase dihydrodipicolinate synthase conserved hypothetical protein aspartate kinase conserved hypothetical protein possible MesJ homolog conserved hypothetical protein excinuclease ABC subunit B putative DNA mismatch repair protein Putative 6,7-dimethyl-8-ribityllumazine synthase o preprotein translocase SecA subunit Online Supporting Information ftsY argH grpE dnaJ murB murC gap2 thiL efp accB pdxA cbiD guaA thiG ycf37 rpl20 rpl35 dnaX clpX asd dapA lysC uvrB mutS ribH secA Table 2 Pag ort327 ort189 ort420 ort445 ort397 ort263 ort009 ort641 ort438 ort638 ort627 ort296 ort645 3.4146 5.5755 2.6576 6.7596 2.1798 8.6248 7.0173 2.6652 4.3220 3.5393 4.3404 4.0367 1.2772 1 1 1 0 1 0 0 1 0 1 0 1 1 all4037 all1076 all4623 alr5266 all5265 alr0517 alr1414 alr0088 all4071 alr5070 all2894 all2102 all0982 glr2514 glr4252 gll1756 gll3248 gll2506 gll0502 glr3183 glr2942 gll3356 gll1494 gll3119 glr2540 glr1118 1638 1637 1636 1635 1634 1627 1625 1623 1618 1616 1615 1613 1611 0117 0118 0119 0120 0121 0136 0138 0141 0147 0155 0156 0158 0160 479.50 448.6 483.54 483.83 483.40 454.52 476.44 444.42 507.275 418.38 431.9 389.22 504.76 125.197 136.2611 136.2498 129.624 129.537 136.2624 127.357 136.2723 125.172 135.2092 132.1024 135.1997 134.1422 1800 1799 1798 1797 1796 1788 1786 1784 1779 1776 1775 1773 1771 slr1348 sll1961 slr0974 sll0817 sll2005 slr1448 sll1234 slr0925 slr1550 slr1639 sll0613 sll0100 slr0118 tlr0851 tll2117 tlr1620 tlr0648 tll0647 tll1428 tll2390 tlr1490 tll0212 tll1927 tll2253 tll0013 tlr0334 99.8760 105.623 114.2168 120.4905 120.4740 121.6195 122.7559 101.106 115.2623 115.2649 112.1715 119.4412 105.559 0090 0091 0093 0094 0095 0117 0119 0121 0127 0134 0135 0137 0140 232.652 232.2627 232.1009 232.1471 232.1470 232.1845 232.2846 232.5313 232.2062 232.1286 232.4875 232.4927 232.4245 354.4128 286.2198 362.6249 361.5471 362.6726 52.32 360.5181 351.3997 362.6074 361.5795 289.2256 361.5413 361.5939 ort652 ort349 ort005 ort518 ort380 ort566 ort091 ort093 ort411 ort294 ort234 ort526 ort528 ort524 ort523 ort288 ort626 ort468 ort474 ort124 ort218 ort216 ort298 ort358 ort335 ort135 ort351 ort406 ort139 ort211 3.4278 3.4950 3.9182 0.3821 4.1998 6.0249 11.8245 7.1695 3.8460 3.9272 10.7885 3.3219 1.9807 2.1270 3.1238 5.6687 4.4346 1.9352 4.2351 4.5468 6.1874 10.6923 3.0127 8.3583 6.2350 5.5160 2.9287 2.6944 7.0841 6.2244 1 1 1 1 1 0 0 0 1 1 0 1 0 1 1 0 1 1 1 1 1 0 1 0 1 0 1 1 0 0 alr3344 alr3343 asr3342 asr3463 alr3464 alr2812 alr2709 alr3454 all1897 all4495 all3989 asr3848 asr3847 asr3846 asr3845 alr3844 alr3843 all3842 all3840 alr2298 alr0180 alr0181 all0278 asr2953 alr3012 all0513 all4391 all4390 all5088 all5089 glr2297 gll4014 glr2311 gsl3287 gll2215 gll1505 gll2564 gll0917 gll0410 glr2394 gll1453 gsr0859 gsr0858 gsr0857 gsr0856 glr0855 glr0854 glr0748 glr0750 glr4164 glr4114 glr4115 glr0870 glr2647 glr1861 glr2108 glr4188 gll0769 glr0532 glr0388 1610 1609 1608 1607 1606 1605 1598 1597 1594 0304 0303 0300 0299 0298 0297 0296 0295 0294 0292 0291 0290 0289 0288 0287 0284 0283 0282 0281 0279 0278 1957 1956 1955 1954 1953 1951 1942 1938 1685 1917 1910 1899 1898 1897 1896 1895 1894 1893 1891 1890 1889 1888 1887 1886 1884 1883 1881 1880 1872 1871 373.12 373.11 373.9 507.20 507.21 443.69 493.69 504.121 400.25 463.56 509.252 493.54 493.53 493.52 493.51 493.50 493.49 493.109 493.111 481.60 495.86 495.87 480.38 482.47 439.69 397.10 493.129 493.131 475.25 475.24 136.2842 136.2841 136.2840 136.2232 136.2233 131.788 132.1078 129.614 135.1786 130.671 134.1487 134.1370 134.1369 134.1368 134.1367 134.1366 134.1365 134.1603 134.1605 136.2771 136.2751 136.2752 131.792 131.923 136.2493 135.1719 136.2459 136.2460 128.515 128.513 1770 1769 1768 1767 1766 1764 1756 1753 1750 0335 0334 0331 0330 0329 0328 0327 0326 0325 0323 0322 0321 0320 0319 0318 0316 0315 0314 0313 0311 0310 sll1070 sll1069 ssl2084 ssl0563 sll0220 slr0808 slr1438 slr1342 sll1184 sll1143 sll1253 smr0008 smr0007 smr0006 ssr3451 slr2034 slr2033 slr1279 slr1281 sll0154 sll1001 sll1002 slr1777 slr1093 slr1666 sll1289 slr1051 slr0500 slr0402 slr1334 tll1870 tll1871 tsl1872 tsl1013 tll1237 tlr1065 tll1673 tll2430 tll0365 tlr2459 tll0519 tsr1544 tsr1543 tsr1542 tsr1541 tll1695 tll1696 tlr1429 tlr1430 tlr1951 tll0328 tll2407 tll1413 tll0178 tlr0710 tlr1788 tll1693 tll0133 tll1797 tlr1520 122.6452 122.6451 122.6450 122.7684 122.7687 122.7168 120.4906 111.1637 116.2796 104.450 103.378 106.653 106.652 106.651 106.650 106.649 106.648 106.714 106.716 109.1102 109.1205 109.1206 102.249 118.3672 93.8356 122.7432 87.8161 87.8165 122.7783 119.4338 0141 0142 0143 0144 0145 0149 0160 0162 0171 0181 0188 0201 0202 0203 0204 0205 0206 0207 0209 0210 0211 0212 0213 0216 0220 0221 0225 0226 0234 0235 232.2543 232.2544 232.2545 232.2386 232.2387 232.4959 232.3146 232.2378 232.2660 232.2265 232.690 232.5532 232.829 232.830 232.831 232.832 232.833 232.834 232.836 232.4018 232.1612 232.1613 232.2001 232.4816 232.4767 232.1841 232.2190 232.2191 232.1328 232.1327 361.5431 362.6015 362.6014 361.5974 362.6625 301.2515 342.3654 361.5829 359.4859 207.1070 358.4650 86.164 86.163 86.162 86.161 86.160 86.159 174.729 174.731 296.2411 343.3660 343.3661 238.1440 274.1977 216.1168 361.5903 186.837 282.2133 349.3910 322.3044 Online Supporting Information cysE infC miaA gyrB ahcY ssb lysS smpB ruvB thiC cbbT, tktA fabF acpP psaC glmS rimM ho1 psbJ psbL psbF psbE ycf48 rub ndhC ndhJ chlD folK degT fabI hisB serine acetyltransferase conserved hypothetical protein translation initiation factor IF-3 tRNA delta-2-isopentenylpyrophosphate (IPP) transfe DNA gyrase subunit B Putative fructokinase putative adenosylhomocysteinase single-stranded DNA-binding protein lysyl-tRNA synthetase tmRNA binding protein SmpB Holliday junction DNA helicase RuvB Zinc metallopeptidase M20/M25/M40 family thiamin biosynthesis protein transketolase 3-oxoacyl-[acyl-carrier-protein] synthase II acyl carrier protein (ACP) photosystem I iron-sulfur center subunit VII (PsaC Glutamine--fructose-6-phosphate transaminase (is possible 16S rRNA processing protein RimM conserved hypothetical protein conserved hypothetical protein Heme oxygenase UvrD/REP helicase possible polyA polymerase putative photosystem II reaction center J protein photosystem II reaction center L protein (psII 5 Kd pr Cytochrome b559 beta chain cytochrome b559 alpha chain similar to plant photosystem II stability/assembly fact probable rubredoxin NADH dehydrogenase I chain 3 (or A) NADH dehydrogenase I chain J conserved hypothetical protein possible ABC transporter, ATP-binding componen possible ABC transporter Protoporphyrin IX Magnesium chelatase subunit C possible 2-amino-4-hydroxy-6-hydroxymethyldihydro putative pleiotropic regulatory protein conserved hypothetical protein enoyl-[acyl-carrier-protein] reductase imidazoleglycerol-phosphate dehydratase conserved hypothetical protein Phosphoglucomutase/phosphomannomutase family p Table 2 ort547 ort247 ort527 ort644 ort320 ort142 ort243 ort439 ort290 ort143 ort388 ort063 ort427 ort144 ort529 ort525 ort531 ort146 ort147 ort148 ort475 ort666 ort414 ort372 ort545 ort336 ort530 ort326 ort022 ort150 ort054 ort053 ort055 ort201 ort295 ort003 ort151 ort253 ort533 ort383 ort152 ort679 ort048 ort363 Pag 6.8194 3.9718 2.5918 3.8372 8.6386 2.6218 10.1699 3.4945 5.6424 7.2182 2.8950 5.8566 9.6463 10.3707 4.9395 3.1472 6.4828 5.8805 8.2031 5.1328 2.6719 8.6804 4.1862 6.4063 4.2890 7.3614 6.1353 13.9143 5.1566 6.6164 1.9188 3.4352 9.8770 4.1417 4.9136 2.7860 6.7278 8.3556 3.5702 1.9370 5.6983 7.5593 6.8278 7.0142 0 1 0 1 0 0 0 1 0 0 1 0 0 0 1 1 0 0 0 0 1 0 1 1 0 0 0 0 0 0 1 1 0 1 0 1 0 0 1 1 0 0 0 0 alr5099 all5094 asl0885 alr1885 alr0379 asl3981 all3113 all0985 alr3085 all0185 alr4806 alr4808 all1416 alr1278 asr0847 asl0846 all0844 asl4395 alr4394 alr4393 alr4392 alr0570 alr1073 asl3192 all1681 all3111 all3854 all5138 all0797 all2705 alr2492 alr2493 alr2494 all4087 asl0940 alr0939 alr4005 alr1128 alr3395 all2319 all1162 alr3123 alr3122 all0154 gll4276 glr1860 gsr2807 glr0802 gll3727 gll0064 gll3761 glr3299 gll1734 gll1471 glr4369 glr2441 glr3418 gll3632 gsl3001 gsr3002 gll0538 gsr0107 gll2308 gll2309 gll3285 glr1718 gll3651 gsl3282 gll2875 glr1548 glr3691 glr1084 glr3393 glr4150 glr1370 glr1371 glr1372 gll0929 gsr2010 gll2012 glr2712 glr3322 glr0043 gll0256 gll3593 glr2021 glr1719 gll3581 Online Supporting Information 0275 0273 0272 0271 0270 0268 0267 0264 0262 0261 0258 0257 0255 0254 0252 0251 0250 0249 0248 0247 0246 0242 0238 0235 0233 0229 0228 0227 0224 0075 0073 0072 0071 0067 0061 0060 0059 0057 1465 1463 1461 1460 1459 1458 1868 1865 1863 1862 1861 1859 1858 1854 1852 1851 1847 1845 1843 1841 1838 1837 1835 1834 1833 1832 1831 1826 1817 1815 1807 1802 1800 1799 1795 1608 1606 1605 1604 1596 1589 1588 1587 1584 1484 1481 1478 1477 1476 1475 475.89 475.17 509.247 509.246 509.116 507.159 489.110 490.102 397.16 492.138 356.2 356.3 459.4 492.17 501.138 501.108 501.110 493.128 493.20 493.19 493.18 509.177 406.27 387.11 378.7 489.112 502.159 458.5 466.46 388.13 502.169 502.171 502.172 494.47 501.60 501.176 504.190 479.56 397.35 475.84 501.196 469.13 469.12 502.44 136.2543 136.2563 136.2807 136.2806 136.2284 136.2285 136.2804 136.2491 135.1953 135.1872 136.2375 136.2377 129.560 135.1701 136.2405 136.2679 136.2681 136.2682 136.2629 136.2628 136.2627 130.771 125.157 131.821 127.391 136.2717 136.2719 136.2720 136.2674 126.256 135.1721 135.1722 135.1723 133.1220 135.1861 133.1255 133.1225 135.1948 136.2360 136.2358 135.2064 136.2357 136.2356 136.2355 0307 0305 0304 0303 0302 0300 0299 0296 0294 0293 0290 0289 0287 0286 0284 0283 0281 0280 0279 0278 0277 0272 0268 0265 0262 0258 0257 0256 0253 0088 0086 0085 0084 0080 0075 0074 0073 0071 1619 1616 1614 1613 1612 1611 slr0185 slr0338 sml0005 slr0713 slr0636 sml0011 sll1284 slr0348 sll2014 slr0488 sll1931 slr0427 sll1444 sll0350 smr0009 ssl2598 slr0922 sll1340 sll0832 sll1424 sll1423 sll1980 sll1362 slr0033 slr1476 sll0250 sll0427 slr1165 sll1747 sll0185 slr0074 slr0075 slr0076 slr1234 ssl0353 sll0053 slr1677 sll1120 sll0421 ssl0707 slr0711 slr2087 sll0621 slr1267 tlr1828 tll1731 tsl0176 tll0080 tll1337 tsr1779 tlr2038 tlr1041 tlr1697 tll2350 tlr2127 tlr2129 tlr1234 tlr1075 tsr1387 tsl1386 tll1384 tsl2214 tlr0651 tll1784 tll1650 tlr0355 tlr2330 tsl1524 tll1558 tll2467 tll0444 tll1044 tll0463 tll2067 tll0490 tlr1904 tlr1905 tlr0997 tlr1577 tlr1808 tll1550 tlr1925 tll1829 tll0591 tll0218 tll0683 tlr0052 tlr0440 105.603 110.1335 120.4838 105.566 105.600 120.4861 122.6578 121.5850 122.6657 122.7476 122.6602 122.6605 108.1038 121.5706 104.494 104.396 104.398 104.402 87.8177 87.8176 87.8174 122.6363 86.8139 122.7025 117.3410 122.6581 120.4767 122.7556 104.495 122.7320 117.3395 117.3396 117.3397 107.845 121.5436 121.5934 121.5738 122.6323 120.4569 121.6103 121.5909 118.3934 118.3933 122.6816 0238 0240 0243 0244 0245 0247 0248 0252 0254 0255 0259 0261 0263 0265 0268 0269 0271 0272 0273 0274 0275 0280 0289 0292 0296 0301 0303 0304 0308 0313 0319 0320 0321 0328 0335 0336 0337 0341 0405 0462 0463 0471 0472 0475 232.1318 232.1323 232.3352 232.2648 232.1757 232.701 232.2703 232.1875 232.3711 232.1616 232.1047 232.1049 232.2848 232.1923 232.3317 232.3316 232.3314 232.2186 232.2187 232.2188 232.2189 232.4228 232.2666 232.2776 232.5037 232.2701 232.820 232.1341 232.4469 232.3142 232.4311 232.4312 232.4313 232.4692 232.4400 232.4399 232.682 232.3545 232.2313 232.4038 232.3283 232.2714 232.2713 232.5371 357.4370 289.2251 361.5328 362.6159 256.1697 321.2998 304.2597 360.4968 359.4836 345.3723 360.5077 362.6743 216.1169 362.6423 319.2958 305.2618 214.1136 332.3321 186.836 186.835 186.834 361.5842 360.4887 253.1647 362.6596 192.895 359.4858 259.1746 280.2085 275.2006 264.1818 264.1819 362.6578 361.5735 333.3352 328.3194 362.6426 282.2136 355.4168 361.5924 321.3011 355.4178 355.4177 360.5218 pyrE psbK tgt cobS lytB glyA leuD psbN psbH pth ntcA txlA ileS gatC pyrB dfp psbO cysD aroC accC purB glnB ycf44 ccdA ftsW Orotate phosphoribosyltransferase probable oxidoreductase photosystem II 4 Kda protein psbK precursor tRNA-guanine transglycosylase Cobalamin-5-phosphate synthase CobS conserved hypothetical protein probable esterase LytB protein homolog sugar fermentation stimulation protein conserved hypothetical protein serine hydroxymethyltransferase (SHMT) CinA-like protein 3-isopropylmalate dehydratase small subunit conserved hypothetical protein Photosystem II reaction centre N protein (psbN) Photosystem II 10 kDa phosphoprotein (PsbH) putative peptidyl-tRNA hydrolase (PTH) conserved hypothetical protein conserved hypothetical protein conserved hypothetical protein Global nitrogen regulatory protein, CRP family of thioredoxin-like protein TxlA isoleucyl-tRNA synthetase glutamyl-tRNA (Gln) amidotransferase subunit C aspartate carbamoyltransferase putative p-pantothenate cysteine ligase… photosystem II manganese-stabilizing polypeptide ATP-sulfurylase Chorismate synthase conserved hypothetical protein ABC transporter, membrane component ABC transporter, ATP-binding component ABC transporter, membrane component HIT (Histidine triad) family protein YGGT family, conserved hypothetical integral memb acetyl-CoA carboxylase, biotin carboxylase subuni conserved hypothetical protein putative chromosome segregation protein, SMC ATPa Fumarate lyase:Adenylosuccinate lyase nitrogen regulatory protein P-II conserved hypothetical protein putative c-type cytochrome biogenesis protein Ccs1 putative c-type cytochrome biogenesis protein CcdA Cell division protein FtsW Table 2 ort200 ort029 ort038 ort037 ort036 ort035 ort033 ort030 ort032 ort153 ort464 ort034 ort031 ort154 ort495 ort550 ort261 ort605 ort155 ort454 ort157 ort634 ort512 ort158 ort662 ort590 ort586 ort465 ort535 ort534 ort434 ort352 ort432 ort433 ort204 ort159 ort669 ort548 ort065 ort160 ort268 ort419 ort161 ort476 Pag 5.2920 8.5178 2.8367 1.1663 6.3709 8.4030 7.3539 2.2227 3.6133 4.6848 6.1496 4.6828 1.4946 3.7880 5.2701 5.3987 7.8110 5.1082 9.8678 7.4248 7.7086 2.9645 9.2621 4.0746 7.2425 2.5600 2.5480 4.3429 6.2194 5.9641 6.1640 3.3057 4.9804 6.6264 5.2804 2.3518 3.8947 7.6277 5.8252 8.7134 7.6197 3.1253 7.4376 2.4787 0 0 1 1 0 0 0 1 1 1 0 1 1 1 1 1 0 1 0 0 0 1 0 1 0 1 1 1 0 0 1 1 1 0 0 1 1 0 1 0 0 1 0 1 all0906 all0011 all0010 asl0009 all0008 all0007 all0006 all0005 all0004 alr0786 alr2485 all5038 all5039 asl4181 all4182 alr2083 alr1264 asr4648 alr4650 alr4112 alr2890 alr1890 alr1891 alr5152 alr1543 asl0146 all0147 alr3511 alr3510 alr2268 alr2270 alr2271 alr2272 alr2274 alr0237 alr2980 alr2982 alr2983 all0492 alr4016 alr3705 alr3832 asr3830 alr3829 gll3181 gll2911 gll2910 gsl2909 gll2908 gll2907 gll2906 gll2905 glr4315 gll0557 glr2587 gll2568 gll2570 glr0831 gll0756 glr0827 gll1676 gsl1620 glr1399 gll3732 gll0391 glr2139 glr2136 glr4119 gll3690 gsl0824 gll0825 gll0055 glr2779 glr1916 glr1865 glr1866 glr1867 glr1868 glr2611 glr0782 gll4415 gll1658 gll0663 gll0664 gll4250 glr2652 glr2651 glr3489 Online Supporting Information 1464 1457 1456 1455 1454 1453 1452 1451 1450 1447 1446 1439 1438 1435 1434 1433 1432 1431 1430 1364 1356 1354 1353 1349 1347 1345 1344 1341 1340 1339 1337 1336 1335 1334 1332 1330 1329 1328 1327 1325 1323 1494 1493 1492 1482 1473 1472 1471 1470 1469 1468 1467 1466 1463 1461 1452 1451 1447 1446 1445 1444 1442 1441 1434 1433 1431 1430 1426 1422 1421 1420 1417 1416 1414 1412 1411 1410 1409 1407 1404 1403 1402 1401 0478 1398 1528 1527 1526 372.18 502.192 502.193 502.194 502.195 502.196 502.197 502.198 502.199 478.98 407.12 431.32 431.30 366.9 366.8 388.8 474.52 489.4 489.6 507.171 420.37 507.265 507.266 507.244 477.121 457.24 457.23 504.61 494.77 440.13 440.16 440.17 440.18 440.19 506.152 423.49 423.51 423.52 412.49 412.47 473.95 479.34 479.33 479.32 136.2732 136.2735 136.2736 136.2737 136.2738 136.2739 136.2740 136.2741 136.2742 136.2744 136.2470 131.816 131.815 136.2274 136.2273 128.450 136.2620 135.1706 135.1708 135.1675 133.1324 133.1160 133.1161 135.1971 124.129 124.128 124.127 126.282 126.281 126.231 126.233 126.234 126.235 126.236 124.111 129.572 129.573 129.574 135.1879 136.2769 136.2217 135.1965 135.1964 135.1963 1617 1610 1609 1608 1607 1606 1605 1604 1603 1601 1599 1592 1591 1588 1587 1586 1585 1584 1583 1439 1438 1436 1435 1430 1428 1426 1425 1422 1421 1420 1418 1417 1416 1415 1413 1411 1410 1409 1408 1406 1404 1649 1648 1647 slr0950 sll1321 sll1322 ssl2615 sll1323 sll1324 sll1325 sll1326 sll1327 slr1287 slr1691 slr1330 slr1329 ssr3307 slr1945 sll0368 slr0086 ssl2982 slr0249 sll2010 sll1252 sll1908 sll1909 slr0050 slr0457 ssr2799 slr1678 sll0698 slr1159 slr1226 sll1508 sll1605 sll0379 slr0015 sll2001 slr1915 slr1031 sll0838 sll1973 slr0730 sll1374 slr0744 ssr1238 slr0743 tll1457 tlr0429 tlr0430 tlr0431 tlr0432 tlr0433 tlr0434 tlr0435 tll0385 tlr0672 tlr1180 tlr0526 tlr0525 tsl1100 tlr0151 tll2320 tll2215 tsr2257 tlr1446 tll0073 tlr2161 tlr0325 tlr0326 tll0915 tlr1606 tsl0166 tll0167 tlr0437 tlr1839 tll1496 tlr1790 tlr1791 tlr1792 tll0321 tlr1745 tll2044 tlr0050 tll0395 tll1107 tll1109 tlr1521 tlr1066 tsl1936 tll1937 122.6696 122.6792 122.6793 122.6794 122.6795 122.6796 122.6797 122.6798 122.6801 100.51 119.4282 113.2071 113.2070 121.5186 121.5185 119.4313 114.2287 121.6012 121.6014 121.5853 120.5114 120.4551 120.4552 118.3927 112.1867 121.5643 121.5642 103.322 103.320 122.6948 113.2015 113.2016 113.2017 113.2020 96.8550 122.7672 122.7673 122.7675 119.4099 119.4097 122.6726 120.4723 120.4722 120.4721 0482 0488 0489 0490 0491 0492 0493 0494 0495 0499 0502 0511 0512 0518 0519 0520 0523 0525 0526 0530 0531 0533 0534 0538 0544 0546 0547 0551 0552 0554 0556 0557 0558 0559 0562 0567 0568 0571 0577 0581 0583 0597 0599 0600 232.3375 232.1561 232.1560 232.1559 232.1558 232.1557 232.1556 232.1555 232.1554 232.4447 232.4304 232.1258 232.1259 232.4601 232.4600 232.2042 232.4453 232.986 232.984 232.4668 232.4879 232.2652 232.2653 232.1359 232.3824 232.5365 232.5366 232.2136 232.2137 232.3994 232.3996 232.3997 232.3998 232.3999 232.1665 232.4794 232.4792 232.4791 232.1834 232.670 232.2482 232.843 232.844 232.845 114.290 362.6283 362.6284 362.6285 362.6286 362.6287 362.6288 362.6289 362.6290 211.1101 362.6471 280.2097 280.2096 277.2029 277.2028 362.6742 360.5052 188.857 188.859 312.2770 309.2703 362.6395 362.6396 276.2011 362.6585 345.3730 345.3729 197.946 360.4899 323.3060 323.3062 310.2718 350.3971 350.3972 315.2839 305.2610 362.6746 344.3688 316.2864 316.2862 320.2981 309.2700 309.2699 309.2698 atp1 atpI atpH atpG atpF atpD atpA atpC nadE atpE atpB pgmI pyrR rpoZ murD serA prmA truB rpl27 rpl21 nblS purD purC lpxC fabZ lpxA lpxB tyrS pyrF infB nusA hemolysin-like protein possible ATP synthase protein 1 ATP synthase subunit A ATP synthase subunit c putative ATP synthase subunit B' putative ATP synthase B chain putative ATP synthase delta chain ATP synthase subunit alpha ATP synthase subunit gamma conserved hypothetical protein putative glutamine dependent NAD(+) synthetase putative ATP synthase epsilon subunit ATP synthase beta subunit conserved hypothetical protein Phosphoglycerate mutase, co-factor-independent ( possible pyrimidine operon regulatory protein Pyr putative DnaK-type molecular chaperone (HSP70 fam putative DNA-directed RNA polymerase (omega c conserved hypothetical protein UDP-N-acetylmuramoylalanine--D-glutamate ligase conserved hypothetical protein putative D-3-phosphoglycerate dehydrogenase (PG putative methyltransferase for Ribosomal protein L11 conserved hypothetical protein putative tRNA pseudouridine 55 synthase 50S ribosomal protein L27 50S ribosomal protein L21 two-component sensor histidine kinase phosphoribosylglycinamide synthetase SAICAR synthetase UDP-3-0-acyl N-acetylglucosamine deacetylase (3R)-hydroxymyristoyl-[acyl carrier protein] dehydra UDP-N-acetylglucosamine acyltransferase Lipid-A-disaccharide synthetase Leucine aminopeptidase conserved hypothetical protein t-RNA synthetase, class Ib:Tyrosyl-tRNA syntheta orotidine 5'-phosphate decarboxylase conserved hypothetical membrane protein conserved hypothetical protein putative GPH family sugar transporter translation initiation factor IF-2 conserved hypothetical protein N utilization substance protein A Table 2 ort162 ort574 ort623 ort221 ort603 ort604 ort163 ort164 ort165 ort166 ort215 ort378 ort259 ort366 ort315 ort167 ort269 ort168 ort169 ort171 ort514 ort244 ort235 ort256 ort435 ort430 ort172 ort257 ort551 ort522 ort678 ort417 ort677 ort418 ort345 ort329 ort504 ort245 ort649 ort173 ort628 ort301 Pag 8.1709 5.3387 5.5292 5.3360 1.5346 4.5380 4.5235 6.7162 8.9004 5.6156 5.4518 8.6143 5.8338 8.4453 10.6937 5.6269 3.7840 5.6993 8.1606 1.9577 7.5842 4.0773 7.3444 9.2918 3.4978 6.5219 6.2089 3.0754 5.9182 1.9365 5.2305 2.8518 5.6368 4.7731 3.2475 4.7620 5.6864 3.2275 5.4471 8.3490 9.8279 2.8155 0 0 1 0 1 1 0 0 0 0 1 0 0 0 0 1 1 0 0 1 0 1 0 0 1 0 1 1 0 1 0 1 0 0 1 1 1 1 0 0 1 1 alr3828 all0888 asr1592 alr1593 alr1594 alr1596 all0355 alr4537 asr0043 alr0044 alr0045 alr3182 alr0489 alr3724 all0455 all0876 alr0483 asr0485 alr0486 alr0487 alr0488 alr4178 alr4175 alr4174 all3184 alr3185 alr3603 alr3606 alr1798 alr4291 all4289 alr4627 all4752 asl4195 alr4351 all1092 alr1254 all0723 all4120 all4825 all2297 all0152 glr3488 gll0030 gll0160 gll2367 glr2283 glr4278 gll2079 gll3007 gsl2882 gll2881 gll2880 glr0708 gll2595 glr1457 gll0377 gll4186 gll3210 gsr0474 glr0702 glr0703 glr0704 gll0082 gll0084 gll3538 glr0786 gll4414 glr1484 gll2569 glr3587 glr2324 glr0079 gll1136 glr3257 gsr0413 gll2252 glr0851 gll0636 gll3233 gll4295 glr0759 gll3300 glr1714 Online Supporting Information 1491 1489 1487 1486 1485 1483 1481 1480 1479 1478 1477 1475 1467 1466 0389 0390 0391 0393 0394 0395 0396 0397 0398 0399 0400 0401 0403 0405 0406 1158 1156 1154 1152 1151 1142 1141 1140 1138 1051 1053 1054 1055 1525 1511 1509 1508 1507 1505 1503 1501 1500 1499 1498 1496 1487 1485 0198 0202 0203 0205 0206 0207 0208 0209 0211 0212 0213 0214 0216 0220 0223 1180 1178 1176 1173 1169 1161 1159 1157 1155 1143 1138 1136 1135 479.31 477.48 498.42 498.43 498.45 498.47 452.61 475.111 506.99 506.100 506.101 458.52 468.88 508.101 461.19 463.41 468.80 468.83 468.84 468.86 468.87 366.14 366.12 353.8 420.44 420.14 344.2 344.3 489.32 496.2 504.113 478.2 505.118 374.33 501.168 452.54 429.46 483.89 474.32 484.89 481.38 506.268 135.1962 136.2210 133.1331 133.1332 133.1333 133.1335 135.2105 135.2108 135.2109 135.2110 135.2111 136.2702 128.481 134.1466 135.1782 136.2712 131.831 135.1936 135.1937 135.1938 135.1939 134.1494 134.1492 134.1491 131.882 131.840 126.228 134.1548 131.837 127.382 127.321 125.201 125.164 129.543 133.1327 135.2148 136.2425 136.2854 135.1892 128.440 132.1098 136.2868 1646 1644 1642 1641 1640 1638 1636 1634 1633 1632 1631 1629 1621 1620 0386 0387 0388 0390 0391 0392 0393 0394 0395 0396 0397 0398 0400 0401 0402 1255 1253 1251 1248 1246 1236 1235 1234 1232 1150 1148 1147 1146 slr0742 slr0194 ssl2233 sll1786 sll1787 sll1789 sll0098 slr1098 ssl1263 sll0661 sll0662 slr0072 slr0451 slr0018 slr1879 slr1896 slr1974 ssl0105 slr0556 slr2073 slr0661 slr1508 slr0181 sll1776 sll0947 slr0994 slr1600 sll1841 sll0467 sll0851 sll0226 sll0065 slr0399 ssl3441 sll0019 slr0958 slr0707 sll0245 sll0455 sll1643 sll0896 slr1030 tll1938 tll1273 tsl1188 tll1187 tll0642 tll0640 tll2425 tll0329 tsr2138 tlr2139 tlr2140 tll1704 tlr0350 tll1534 tlr1449 tlr1934 tlr0878 tsl1609 tll1037 tlr1216 tlr1217 tll0327 tlr2234 tlr1079 tlr0833 tll0832 tll1300 tll1299 tll0922 tlr1631 tll1388 tll0880 tll0859 tsr0101 tlr1040 tlr1827 tll1339 tll1587 tll0277 tll0980 tlr1137 tll1511 120.4719 93.8368 100.36 100.37 100.38 100.40 100.17 121.5533 122.6509 122.6510 122.6511 117.3500 100.53 112.1819 118.3831 104.409 116.2932 122.6868 122.6870 122.6874 122.6875 122.7159 122.7155 122.7154 95.8470 95.8463 115.2573 115.2574 118.3847 122.7730 122.6413 115.2593 95.8445 99.8787 116.3018 120.4696 122.7460 121.5444 122.7038 119.4298 119.4463 119.4459 0601 0609 0611 0612 0613 0615 0617 0621 0622 0623 0624 0626 0636 0637 0653 0656 0657 0659 0660 0661 0662 0663 0665 0666 0667 0668 0670 0671 0672 0676 0678 0680 0683 0686 0698 0701 0702 0704 0711 0714 0715 0716 232.846 232.3355 232.3075 232.3076 232.3077 232.3079 232.1739 232.5257 232.1586 232.1587 232.1588 232.2767 232.1829 232.2455 232.1797 232.3344 232.1824 232.5570 232.1826 232.1827 232.1828 232.4604 232.4606 232.4607 232.2769 232.2770 232.2085 232.2087 232.3656 232.5100 232.5098 232.1006 232.894 232.4587 232.5157 232.2612 232.4462 232.3483 232.4658 232.1066 232.4017 232.5369 309.2697 265.1827 350.3956 350.3957 350.3958 350.3959 361.5814 357.4428 291.2286 291.2287 291.2288 330.3242 361.5896 206.1050 362.6590 82.141 356.4254 361.5817 361.5818 359.4834 359.4835 338.3506 202.1004 282.2125 362.6160 362.6612 361.5438 335.3423 300.2486 83.143 362.6489 359.4703 69.88 296.2398 358.4480 356.4250 332.3308 336.3426 362.6617 103.241 361.5674 362.6292 conserved hypothetical protein cbbI, rpiA, ppi rpsT rpoB rpoC2 ycf35 gidB fumC cobI proC lrtA lipB queA psbC ycf4 ilvN ycf39 infA dxr cysRS polA thrA ruvC chlI putative ribose 5-phosphate isomerase A 30S ribosomal protein S20 possible deoxyribonuclease similar to TatD DNA-directed RNA polymerase beta chain RNA polymerase beta prime subunit conserved hypothetical protein conserved hypothetical protein conserved hypothetical protein conserved hypothetical protein possible 3Fe-4S ferredoxin putative glucose inhibited division protein B putative DNA helicase Fumarate hydratase PRECORRIN-2 C20-METHYLTRANSFERASE conserved hypothetical protein putative GTP-binding protein conserved hypothetical protein conserved hypothetical protein conserved hypothetical protein putative pyrroline-5-carboxylate reductase probable glycosyltransferase possible Recombination protein O (RecO) Putative deoxyribose-phosphate aldolase light repressed protein A homolog putative lipoate-protein ligase B conserved hypothetical protein Putative dihydrolipoamide acetyltransferase comp Queuosine biosynthesis protein photosystem II chlorophyll-binding protein CP43 photosystem I assembly protein (Ycf4 family) Acetolactate synthase small subunit putative chaperon-like protein for quinone binding in translation initiation factor IF-1 1-deoxy-D-xylulose 5-phosphate reductoisomerase cysteinyl-tRNA synthetase DNA polymerase I probable GTP-binding protein homoserine dehydrogenase conserved hypothetical protein Holliday junction resolvase RuvC Protoporphyrin IX Magnesium-chelatase subunit Table 2 Pag ort249 ort330 ort491 ort175 ort409 ort663 ort395 ort396 ort176 ort425 ort357 ort324 ort045 ort682 ort492 ort496 ort405 ort178 ort179 ort180 ort181 ort513 ort426 ort004 6.2356 6.5083 2.2649 8.0009 7.5469 2.0190 1.7450 2.9578 6.3910 2.5893 4.6210 4.4641 6.8123 4.2712 3.5522 8.8654 5.7866 8.2150 7.9997 8.5324 10.3178 6.4096 3.7672 3.6739 0 0 1 0 0 1 1 1 0 1 0 1 0 1 1 0 0 0 0 1 0 0 1 1 alr4542 all1365 asr1366 all1367 all1368 alr0052 alr0051 all0860 alr2450 alr4840 alr0212 alr0213 alr3934 all4019 all4121 all4063 all4506 all2716 alr3100 alr3101 alr3102 alr3103 alr1313 all2364 glr2140 gll2782 gsl0511 gll2930 glr0407 gll0880 glr1603 gll3082 glr4151 gll2290 gll1884 gll0416 glr3695 glr3178 gll2295 glr0301 glr0706 glr0793 gll1132 gll1131 gll1129 gll0069 gll3551 glr1605 1056 1057 1058 1059 1060 1061 1062 1063 1065 1066 1069 1070 1072 1074 1075 0798 0796 0794 0791 0790 0789 0788 0786 0784 1132 1131 1130 1129 1128 1127 1126 1124 1122 1121 1110 1109 1104 1102 1101 0518 0520 0523 0526 0527 0528 0529 0531 0534 348.20 498.50 498.112 498.49 498.48 493.78 493.77 424.29 506.176 505.188 504.198 504.199 505.210 507.156 474.31 508.85 455.50 466.40 509.282 509.283 509.285 509.286 497.43 497.48 135.1633 133.1173 133.1310 133.1172 133.1171 135.2074 135.2073 136.2694 135.2041 133.1210 132.1115 132.1116 135.1784 131.893 134.1488 135.1770 135.1773 129.593 125.183 125.184 125.185 130.682 133.1159 135.1833 1144 1143 1142 1141 1140 1139 1138 1137 1135 1134 1130 1129 1126 1124 1123 0874 0872 0870 0867 0866 0865 0864 0862 0859 slr0120 sll1245 smr0010 slr0383 slr0084 slr0623 slr1722 slr0417 sll0735 slr0186 sll0753 slr0739 sll1501 slr1843 slr1643 sll1522 slr0652 slr1796 sll1547 sll1109 slr0362 slr2035 slr1517 sll0336 tll2116 tll2429 tsr0877 tlr1053 tlr1536 tll0812 tll2190 tlr1369 tll1974 tll1397 tll0021 tll0020 tlr1848 tll0540 tlr1211 tll1275 tll0216 tlr1421 tll2329 tll1867 tll0495 tll2118 tlr1600 tlr1643 120.4823 109.1123 109.1189 109.1121 109.1120 113.2011 113.2003 120.5100 122.7759 122.6760 120.5087 120.5088 122.7733 118.3670 119.4027 116.2896 121.5431 121.5725 117.3230 117.3232 117.3234 110.1319 117.3435 122.6621 0719 0720 0721 0722 0723 0724 0725 0727 0729 0730 0740 0742 0748 0750 0751 0771 0773 0776 0779 0780 0781 0782 0784 0788 232.5260 232.2899 232.5612 232.2898 232.2897 232.1595 232.1594 232.3329 232.4270 232.1082 232.1641 232.1642 232.748 232.666 232.4657 232.628 232.2276 232.3153 232.2694 232.2695 232.2696 232.2697 232.1911 232.4084 310.2722 361.5608 361.5636 339.3528 362.6518 207.1057 359.4808 321.3012 340.3569 276.2012 361.5626 268.1877 76.122 324.3080 343.3658 357.4422 362.6327 212.1113 292.2320 300.2498 71.95 324.3087 361.5260 362.6124 ort044 ort542 ort182 ort332 ort300 ort359 2.1308 4.5633 5.4837 3.6056 2.4003 8.2432 1 0 0 1 1 0 all4563 alr2475 asr2474 alr2542 all4365 alr4386 gll0752 glr1325 gsl0442 gll1206 gll2622 glr0540 0781 0780 0779 0832 0831 0830 0537 0539 0540 0814 0815 0816 493.70 465.67 465.66 498.2 440.25 501.135 133.1293 132.1010 132.1009 130.766 130.649 131.808 0855 0854 0853 0904 0903 0902 sll0018 slr0520 slr0519 sll1058 slr1055 slr2026 tlr0376 tlr2187 tsr2185 tll1987 tlr0271 tlr1522 120.4621 110.1374 110.1373 119.3993 121.5660 112.1727 0791 0793 0794 0819 0820 0822 232.1445 232.4294 232.4293 232.4360 232.2217 232.2195 332.3318 362.6487 362.6486 360.4914 357.4373 205.1038 ort654 ort057 ort183 ort043 ort184 ort185 ort367 ort197 ort061 ort187 ort485 4.8826 6.1935 8.0607 3.3436 6.4854 4.9317 4.8449 3.0645 5.3822 7.2858 4.6257 1 0 0 1 1 1 0 1 0 0 0 alr4385 all0640 alr4979 alr3809 alr4169 alr4171 all1691 alr0799 asr0798 all4869 all0218 gll1040 gll2329 gll3597 gll1769 gsl0787 gll2939 glr3304 glr3340 gsr4417 gll3437 gll0103 0829 0827 1170 0825 0823 0821 0637 1111 1110 1109 1107 0817 0819 0821 0822 0824 0825 0858 1095 1094 1093 1091 501.134 479.58 399.18 479.11 353.3 353.5 463.18 443.28 443.27 443.26 504.10 128.519 136.2606 136.2554 121.7 136.2648 136.2650 134.1479 134.1390 134.1389 124.110 124.118 0901 0899 1181 0897 0895 0893 0794 1085 1086 1087 1089 slr0783 sll1276 slr1900 sll0370 slr1886 sll0585 sll0567 slr1846 ssr3122 slr1160 slr1779 tlr0966 tlr0962 tll0916 tll2332 tlr1222 tll1062 tll0048 tll0874 tsl0875 tll0135 tlr0838 112.1726 120.5139 113.2078 104.406 122.7175 122.7177 110.1378 118.3891 118.3890 118.3889 120.4563 0823 0825 0829 0830 0832 0834 0865 0906 0907 0908 0910 232.2196 232.3432 232.1217 232.868 232.4612 232.4610 232.5028 232.4471 232.4470 232.1108 232.1647 205.1037 321.3002 128.365 361.5742 359.4813 359.4815 360.4935 247.1570 247.1569 224.1263 324.3095 Online Supporting Information cytM petG hisH trxA guaB gyrA leuA folD crtE cbiA zwf petH pgsA hisA proB leuB accD cbbA, cfxA, fbaA, fda purQ dapB chlH folP cbbJ, tpiA carB fur pdxJ probable tRNA/rRNA methyltransferase cytochrome cM Cytochrome b6f complex subunit 5 conserved hypothetical protein Glutamine amidotransferase class-I Thioredoxin putative IMP dehydrogenase DNA gyrase A subunit conserved hypothetical protein 2-isopropylmalate synthase methylenetetrahydrofolate dehydrogenase geranylgeranyl pyrophosphate synthase putative Cobyrinic acid a,c-diamide synthase glucose-6-phosphate dehydrogenase ferredoxin--NADP reductase (FNR) CDP-diacylglycerol-glycerol-3-phosphate 3-phosphat phosphoribosylformimino-5-aminoimidazole carboxa conserved hypothetical protein conserved hypothetical protein conserved hypothetical protein conserved hypothetical protein putative glutamate 5-kinase 3-isopropylmalate dehydrogenase acetyl-CoA carboxylase, beta subunit Fructose-bisphosphate/sedoheptulose-1,7-bisphosp phosphoribosylformylglycinamidine synthase conserved hypothetical protein dihydrodipicolinate reductase Protoporphyrin IX Magnesium chelatase subunit c putative dihydropteroate synthase triosephosphate isomerase ABC transporter, possibly multidrug efflux conserved hypothetical protein carbamoyl phosphate synthetase, large subunit conserved hypothetical protein conserved hypothetical protein Ferric uptake regulator family glutaredoxin-like protein BolA family protein conserved hypothetical protein Pyridoxal phosphate biosynthetic protein PdxJ Table 2 ort049 ort560 ort039 ort671 ort188 ort436 ort282 ort305 ort265 ort280 ort041 ort625 ort440 ort423 ort266 ort456 ort381 ort066 ort020 ort242 ort233 ort458 ort629 ort006 ort056 ort067 ort042 ort068 ort239 ort370 ort339 ort069 ort611 ort340 ort283 ort422 ort631 ort632 ort070 ort382 ort192 ort273 ort071 ort072 Pag 4.8613 2.5127 8.9380 5.3414 5.7140 4.9091 8.8799 0.9607 6.6372 5.4243 9.1914 7.0924 7.6179 2.0589 3.9430 7.9238 6.0105 7.6070 11.5504 5.6520 3.6327 7.6720 4.3042 3.0815 6.0807 7.5864 4.3204 8.0286 5.1483 4.1255 3.3715 8.5950 4.3506 5.6077 6.2624 7.2306 4.1149 6.9207 7.1654 2.7930 6.0516 8.8208 9.2089 5.9052 1 1 0 0 1 1 0 1 0 0 0 0 0 1 0 0 0 0 0 0 1 0 1 1 0 0 1 0 0 1 1 0 1 0 0 0 1 0 0 1 0 0 0 0 all2146 alr4977 alr1921 all2995 all2996 all2997 alr2998 alr2999 alr2764 all3875 all4887 all3873 alr3871 alr4670 alr1879 all0036 alr3921 alr3919 all5019 all2568 alr2001 alr0094 alr0096 all4257 alr4715 asl1962 alr1155 alr1158 alr1159 all1053 all3578 all0748 asl0749 all1323 alr3229 alr3230 all0121 all0120 alr3655 alr2328 all4872 alr4577 alr4576 asl2557 gll4135 glr3499 glr4328 gll0108 gll0109 gll0110 glr2063 glr2064 gll1456 gll1396 gll3487 glr4397 gll3316 gll0901 glr0932 glr2398 glr0443 gll1313 gll1038 gll2974 glr2943 gll1496 gll0753 gll0159 gll3672 gsl3751 gll3543 glr4180 glr4181 gll1528 glr3934 glr1740 gsl4026 gll4083 gll1991 gll0102 glr4166 glr4167 glr2407 glr1052 gll1053 glr0072 glr4030 gsr1914 Online Supporting Information 1099 1097 1093 1092 1091 1090 1089 1088 1087 1085 1084 0658 1081 1080 0609 0610 0611 0612 0613 0614 0615 0617 0618 0619 0954 0953 0951 0949 0948 0946 0945 0944 0943 0939 0933 0932 0929 0928 0923 0920 0918 0915 0914 0744 1080 1077 1056 1057 1058 1059 1060 1061 1062 1065 1066 1069 1071 1074 0403 0401 0400 0399 0398 0396 0395 0393 0392 0389 0655 1037 0653 0651 0650 0648 0647 0646 0645 0641 0621 0620 0617 0616 0607 0601 0599 0595 0594 0593 474.91 505.41 479.62 489.65 489.64 489.62 489.55 489.56 492.34 432.43 506.205 432.44 432.17 422.30 402.16 505.89 460.13 460.10 480.22 441.45 493.79 444.48 436.4 462.4 505.57 466.22 477.2 477.5 477.6 480.85 502.210 472.34 472.33 466.16 502.1 502.2 370.26 370.27 430.13 481.19 443.24 430.2 430.1 494.69 135.1906 133.1251 136.2305 136.2700 136.2699 136.2698 136.2385 136.2386 131.915 136.2320 127.299 134.1463 134.1505 130.641 129.604 135.2066 136.2373 135.1823 136.2402 136.2557 131.841 131.844 131.845 128.472 127.373 131.819 130.776 130.778 130.779 130.781 130.734 131.805 131.804 128.497 136.2363 136.2364 136.2451 136.2452 133.1196 130.757 135.1785 135.2122 135.2121 124.120 1096 1098 1102 1103 1104 1105 1106 1107 1108 1111 1112 1114 1116 1118 1052 1051 1050 1049 1048 1046 1045 1043 1042 1041 0742 0743 0745 0747 0748 0750 0751 0752 0753 0757 0763 0764 0767 0768 0774 1038 1036 1032 1031 1030 slr2019 slr1426 slr1364 sll0506 sll0505 sll0504 slr0853 sll0020 slr1167 slr1369 sll1683 slr0361 sll1676 sll0469 sll0945 slr1351 sll0899 sll0983 slr0444 slr1718 sll0601 slr1746 slr0611 sll0542 slr0615 ssr2551 sll1498 slr0954 slr0955 slr0877 slr0603 sll0933 ssl1784 sll1868 sll0708 sll0711 slr0774 slr0775 sll1968 slr1756 sll1631 slr1366 slr1365 ssl3451 tlr0577 tll2204 tll0661 tlr1763 tlr1762 tlr1761 tll0471 tll0307 tll2051 tll2108 tlr1866 tlr0656 tlr0708 tlr1546 tll0763 tlr1553 tlr0393 tll1622 tlr0343 tll0403 tll0920 tlr0461 tlr1757 tll0887 tlr0655 tsr1432 tll0760 tlr0428 tll1042 tll1003 tll2056 tlr1208 tsr1207 tlr1317 tlr0659 tll0500 tll0203 tll0202 tlr1461 tll1588 tlr0177 tlr2420 tll0901 tsl2428 116.3092 103.384 106.684 98.8680 98.8679 98.8675 98.8733 98.8735 121.6207 112.1671 119.4089 112.1672 112.1842 118.3586 109.1118 122.6421 122.6680 122.6679 122.7271 80.8040 116.2875 121.6057 121.5331 115.2420 117.3434 121.5982 112.1859 112.1862 112.1863 114.2352 119.4212 121.5340 121.5339 118.3845 118.3872 122.6913 97.8587 97.8588 122.7363 122.7439 120.5015 106.687 106.686 104.417 0924 0928 0933 0934 0935 0936 0937 0938 0939 0942 0944 0949 0962 0966 1000 1002 1003 1004 1005 1007 1008 1010 1011 1013 1023 1024 1026 1028 1029 1031 1032 1033 1034 1038 1052 1053 1056 1057 1068 1073 1076 1079 1080 1081 232.1937 232.1215 232.3256 232.4781 232.4780 232.4779 232.4778 232.4777 232.3920 232.796 232.1126 232.798 232.800 232.965 232.3630 232.1578 232.756 232.758 232.1221 232.4381 232.3898 232.5318 232.5319 232.5064 232.927 232.3218 232.3277 232.3279 232.3280 232.2640 232.5452 232.3508 232.3509 232.2681 232.3733 232.3734 232.5340 232.5339 232.2531 232.4048 232.1111 232.1430 232.1431 232.4371 281.2107 359.4760 360.4910 356.4269 356.4268 356.4267 268.1879 361.5366 356.4294 312.2764 268.1880 362.6561 362.6305 337.3476 227.1297 353.4119 362.6644 360.4985 129.373 360.5038 353.4111 360.5050 360.5051 361.5663 329.3227 340.3568 360.5089 301.2517 301.2518 330.3245 358.4486 270.1913 362.6736 260.1755 320.2971 295.2379 352.4038 352.4039 309.2711 361.5271 342.3652 360.4912 360.4911 231.1346 recR bioB uppS lysA clpC cad rsuA malQ kprS murF glmU aroA murI sds acs carA gatA dnaE rps15 dnaG ispE secD secF glnA ABC transporter, multidrug efflux family RecR protein biotin synthase Undecaprenyl pyrophosphate synthetase (UPPS) conserved hypothetical protein diaminopimelate decarboxylase putative ribosomal-protein-alanine acetyltransferase endopeptidase Clp ATP-binding chain C putative glycerol dehydrogenase putative phosphatidate cytidylyltransferase Orn/Lys/Arg decarboxylases family 1 putative pseudouridylate synthase specific to ribosom Putative 4-alpha-glucanotransferase Ribose-phosphate pyrophosphokinase Putative glycogen synthase (glgA) Cytoplasmic peptidoglycan synthetases, N-terminal:C UDP-N-acetylglucosamine pyrophosphorylase conserved hypothetical protein EPSP synthase (3-phosphoshikimate 1-carboxyvinyltr probable 2-phosphosulfolactate phosphatase Possible nitrilase Putative Aspartate and glutamate racemases:Glutamat polyprenyl synthetase; solanesyl diphosphate synth acetyl-coenzyme A synthetase ABC transporter, multidrug efflux family conserved hypothetical protein carbamoyl-phosphate synthase small chain conserved hypothetical protein possible tRNA/rRNA methyltransferase glutamyl-tRNA (Gln) amidotransferase subunit A DNA polymerase III, alpha subunit conserved hypothetical protein 30S ribosomal protein S15 putative DNA primase putative rRNA (adenine-N6,N6)-dimethyltransferase Putative 4-diphosphocytidyl-2C-methyl-D-erythritol k preprotein translocase subunit preprotein translocase subunit conserved hypothetical protein Glutamine synthetase, glutamate--ammonia ligase cytidine/deoxycytidylate deaminase family protein putative lipoprotein signal peptidase conserved hypothetical protein conserved hypothetical protein Table 2 ort482 ort225 ort616 ort664 ort558 ort255 ort637 ort390 ort304 ort667 ort217 ort226 Pag 7.5546 4.3031 2.2408 3.2377 5.5049 7.1035 4.7762 7.9863 2.1261 1.9233 4.7233 4.3761 0 0 1 1 0 0 1 0 1 1 0 1 alr3707 all4793 all4792 all4791 all4789 all4116 alr1348 alr4111 alr0128 all4140 alr4068 all0936 glr2589 gll2797 gll1830 gll1829 gll1278 gll1805 glr1848 glr1646 glr4377 gll1520 glr3264 glr2022 0747 0752 0753 0754 0756 0757 0758 0759 0760 0762 0764 0765 0590 0585 0584 0583 0581 0580 0579 0578 0577 0575 0572 0570 493.86 493.62 493.94 493.95 493.98 506.66 448.44 507.170 504.170 389.23 477.41 501.67 124.87 135.1925 129.552 129.553 134.1552 136.2795 136.2582 136.2522 136.2326 133.1336 136.2245 136.2247 0819 0824 0825 0826 0828 0829 0830 0831 0832 0834 0836 0838 slr0116 sll1664 sll1260 sll1261 slr0020 slr1679 slr0963 slr0220 sll1091 slr1105 slr0251 sll1513 tll2308 tll0992 tll1688 tll1687 tll0735 tll2463 tlr0339 tll2192 tll0150 tlr2283 tll1571 tlr1615 120.5073 114.2275 101.96 101.97 101.99 122.6782 88.8199 121.5852 115.2471 122.7918 122.6747 118.3622 1084 1089 1090 1091 1093 1094 1095 1096 1097 1109 1112 1114 232.2484 232.1035 232.1032 232.1031 232.1029 232.4662 232.3880 232.4669 232.5347 232.4639 232.623 232.4396 311.2748 231.1344 355.4199 355.4200 257.1720 234.1382 329.3229 349.3913 225.1268 361.5888 324.3103 289.2253 ort573 5.0177 1 alr0782 gll3548 0766 0569 464.41 127.350 0839 sll0807 tll2369 122.6539 1115 232.3533 359.4803 ort384 ort398 ort008 ort391 ort251 ort287 ort322 ort510 ort073 ort074 ort075 ort076 ort027 ort373 ort062 ort292 ort354 ort681 ort077 ort594 ort655 ort286 ort321 ort078 ort443 ort248 ort613 ort595 2.9450 4.9077 3.3229 4.5135 9.5890 7.1471 5.6617 3.2001 10.1062 4.6868 11.1679 6.2868 5.8281 2.2365 4.7740 3.9710 6.5069 9.0545 7.3219 3.7495 2.5827 5.8235 8.1273 7.7510 5.2283 8.5050 1.7582 2.0761 1 1 1 1 0 0 0 1 0 0 0 1 0 1 1 1 0 0 0 1 0 0 0 0 0 0 1 1 alr1041 alr1042 all4645 alr5275 alr1602 all2063 all3392 all4379 all3259 alr2014 alr1349 alr1612 alr4853 all2501 all2500 all5215 alr3460 alr1240 alr5129 asl3674 alr4641 alr3352 all3174 all3821 all0233 all4450 asl4451 asl4452 glr3342 glr1218 gll4260 gll1117 glr3180 gll0063 gll0123 glr3159 glr1039 glr1395 gll4381 gll0900 glr2600 gll3622 gll3102 gll3837 glr1831 gll1448 glr0972 gsl0792 gll3158 gll0699 glr0747 gll4313 gll3023 gll1616 gsl3059 gsl3060 0767 0768 0769 0770 0771 0776 0778 0663 0664 0667 0668 0669 0674 0676 0677 0839 0841 0847 0850 0853 0856 0862 0863 0865 0867 0868 0869 0870 0568 0567 0566 0565 0564 0558 0556 0791 0789 0787 0786 0785 0779 0777 0776 0775 0772 0766 0763 0757 0754 0750 0749 0745 0743 0741 0740 0739 443.51 443.53 457.46 506.7 397.26 454.26 400.30 370.7 505.36 458.41 448.46 430.23 450.3 492.100 492.102 493.138 504.126 462.61 435.38 448.15 346.9 346.5 405.44 479.120 506.108 498.149 498.147 498.146 136.2249 136.2250 127.345 136.2594 136.2837 135.2130 132.1090 131.846 136.2857 136.2859 132.1000 136.2387 129.557 121.2 121.3 128.454 135.1910 135.2024 136.2872 134.1472 131.905 135.2052 134.1476 135.2145 134.1587 134.1578 134.1577 134.1576 0840 0841 0842 0843 0844 0849 0851 1029 1028 1025 1024 1023 1018 1015 1014 1000 0997 0988 0985 0981 0978 0975 0974 0972 0970 0969 0968 0967 slr2094 slr1808 slr1176 sll0329 sll1479 sll1035 slr0502 slr1228 slr1918 slr1195 slr0722 sll0875 sll0402 slr2136 slr1751 sll0377 slr0070 sll1910 slr0022 ssr1736 sll0755 slr0992 slr0216 slr0948 slr0649 sll1290 ssr1399 ssr1398 tll1276 tll1738 tlr1287 tlr0576 tlr1076 tlr1764 tll1624 tll2205 tlr2348 tlr0208 tlr1746 tlr2384 tll2357 tlr0996 tll1406 tll0952 tlr1874 tlr1873 tlr0653 tsr1915 tll1454 tlr1793 tll1104 tll0685 tlr0750 tlr0652 tsr2061 tsr2060 119.4322 119.4324 117.3427 115.2577 122.6404 109.1072 108.883 104.464 113.2047 121.5979 88.8200 94.8421 116.2825 121.5491 121.5498 111.1522 111.1641 122.6355 112.1656 118.3703 113.1974 113.1971 121.5389 119.4163 115.2487 121.5958 121.5957 121.5956 1116 1117 1118 1119 1120 1125 1127 1149 1152 1155 1156 1158 1171 1174 1175 1177 1195 1202 1205 1210 1213 1215 1216 1220 1222 1224 1225 1226 232.2593 232.2595 232.991 232.1480 232.3085 232.2026 232.2310 232.2204 232.3759 232.3911 232.3879 232.3093 232.1098 232.4319 232.4318 232.1703 232.2384 232.4430 232.1333 232.2512 232.995 232.2105 232.2759 232.854 232.1661 232.5178 232.5177 232.5176 345.3766 352.4040 221.1219 339.3533 314.2823 326.3147 357.4366 362.6535 305.2620 211.1104 223.1242 255.1691 286.2196 197.949 359.4789 360.4970 205.1044 265.1835 345.3746 233.1376 327.3165 315.2859 313.2790 86.156 361.5541 284.2160 284.2159 284.2158 Online Supporting Information pcyA rps2 tsf recG sir glyS chlP typA cbbE, rpe, ppE cbbF, glpX, fbp hemA agp gnd cobW prfC aspC gcpE fmt zam rpl32 tpx cobU metG rps18 rpl33 ferredoxin-dependent biliverdin reductase possible glycosyltransferase 30S ribosomal protein S2 putative elongation factor EF-Ts ATP-dependent DNA helicase putative d-alanyl-d-alanine dipeptidase Ferredoxin-sulfite reductase putative Glycyl-tRNA synthetase, beta subunit geranylgeranyl hydrogenase tyrosine binding protein possible ABC transporter, ATP-binding component possible heme transporter ribulose-5-phosphate 3-epimerase Fructose-1,6-bisphosphatase/sedoheptulose-1,7-bis Possible glutamyl-tRNA reductase ADP-glucose pyrophosphorylase 6-phosphogluconate dehydrogenase Putative 6-phosphogluconolactonase, Pgl, DevB putative uracil phosphoribosyltransferase putative cobalamin synthesis protein peptide-chain-release factor RF-3 conserved hypothetical protein conserved hypothetical protein conserved hypothetical protein conserved hypothetical protein Aminotransferases class-I GcpE protein carboxyl-terminal processing protease Transcriptional-repair coupling factor putative methionyl-tRNA formyltransferase putative acetazolamide conferring resistance protein Z conserved hypothetical protein 50S ribosomal protein L32 thioredoxin peroxidase putative tRNA/rRNA methyltransferase (SpoU family putative cobinamide kinase conserved hypothetical protein putative methionyl-tRNA synthetase probable ribonuclease II 30S ribosomal protein S18 50S ribosomal protein L33 Table 2 ort499 ort318 ort674 ort309 ort333 ort428 ort493 ort541 ort013 ort561 ort246 ort079 ort490 ort203 ort413 ort591 ort058 ort080 ort081 ort346 ort082 ort064 ort544 ort083 ort538 ort021 ort084 ort085 ort461 ort086 ort262 ort660 ort264 ort650 ort355 ort088 ort379 ort400 ort089 ort410 ort090 ort338 ort276 ort220 Pag 6.5051 5.6170 4.3774 5.0622 7.0569 4.3385 9.9771 6.2372 4.8173 3.9281 4.6797 5.8593 4.6015 5.9978 4.3030 3.5379 5.9866 7.6393 6.8549 2.5721 5.5161 4.7155 3.7078 8.4322 7.3409 11.5504 9.1356 4.6858 4.0298 7.1266 8.2632 3.5967 7.6843 7.2256 9.4604 8.8310 3.4350 3.1599 7.2057 3.5568 9.0366 3.6201 5.1709 5.6858 0 0 1 1 0 1 0 0 0 1 1 0 1 0 1 1 0 0 0 1 0 1 1 0 0 0 0 0 1 0 0 1 0 1 0 0 1 1 0 1 0 1 1 0 alr4958 alr1689 alr2728 alr4700 alr2048 alr3283 alr1050 all0788 alr3537 all3536 alr4054 alr2431 all2919 all2917 alr2323 asl2630 all2556 all4508 asl4507 alr0599 all5166 asl2061 all4008 all0646 alr2742 alr1924 alr3357 all2545 all4673 alr2751 all3970 all1269 alr2973 alr0346 alr3512 alr2762 all0713 all3909 all3908 all3263 alr4810 alr2009 alr1978 alr1965 gll2926 glr3076 gll3592 glr0847 gll0815 gll4081 gll1692 glr1401 glr4010 glr0988 gll2424 glr1664 gll3182 glr1322 glr1814 gsr3838 glr3389 glr1876 gsl4025 gll0194 glr1858 gsr3540 gll3495 glr1400 gll1252 glr2513 gll2384 gll4395 gll0192 gll3484 gll0656 glr3011 gll1169 gll1127 glr1487 gll4416 glr1306 gll3877 glr2049 glr1211 gsr3242 gll1473 gll2868 glr3383 Online Supporting Information 0871 0879 0882 0884 0888 0889 0890 0891 0892 0893 0301 0895 0898 0899 0901 0902 0903 0904 0905 0907 0909 0910 0912 0741 0683 0682 0681 0680 0678 0801 0802 0598 0596 0595 0591 0588 0584 0583 0582 0578 0571 0565 0561 0560 0738 0727 0724 0722 0716 0715 0714 0710 0709 0708 1900 0705 2195 0699 0696 0695 1012 0689 0688 0685 0683 0682 0679 0675 0669 0667 0666 0665 0661 0506 0504 0422 0425 0426 0435 0439 0444 0445 0446 0453 0466 1187 1192 1193 476.101 463.61 452.22 452.25 507.259 505.68 443.58 490.7 484.106 484.30 393.18 489.37 477.92 477.94 484.114 484.115 494.70 455.48 455.49 458.24 498.70 454.28 412.28 378.2 505.195 479.63 498.37 498.172 382.10 506.145 357.12 492.55 492.50 348.5 452.71 492.31 459.48 508.90 508.91 505.33 415.4 415.9 421.26 483.32 128.496 136.2474 135.2008 135.1973 135.1813 135.2019 135.1867 126.262 133.1289 133.1201 126.283 136.2549 136.2552 136.2553 135.2084 136.2514 127.327 133.1148 126.285 136.2793 136.2300 136.2269 136.2473 133.1358 133.1286 136.2238 134.1564 134.1427 133.1295 136.2253 136.2290 128.444 136.2677 133.1194 136.2595 135.2095 134.1559 134.1560 134.1561 135.1979 136.2308 134.1419 132.1111 132.1110 0966 0957 0954 0952 0948 0947 0946 0945 0944 0943 0332 0940 0993 0937 0934 0933 0932 0931 0930 0928 0926 0925 0923 0919 1008 1009 1010 1011 1013 0879 0880 1063 1065 1066 1070 1074 1078 1079 1080 0582 0573 0567 0563 0562 sll1553 slr1211 sll0865 slr0847 slr1665 sll1074 slr1349 slr0477 sll0080 sll1894 sll0135 sll0860 sll1382 sll1383 sll0430 ssr1604 sll0221 sll0608 sll0609 sll1945 slr1577 ssr2142 sll1275 sll1414 sll0578 slr2130 slr0351 sll1071 sll0622 sll1218 slr1822 slr1884 sll0593 sll1760 slr1626 sll1702 sll0158 slr0536 sll0456 slr0608 slr0815 sll0848 slr1149 sll0900 tll0662 tlr0900 tlr1753 tlr1165 tll2294 tll2098 tll0717 tlr2326 tll2219 tlr1727 tlr1281 tll0315 tlr1656 tlr1657 tll1191 tlr2462 tlr1194 tlr1437 tsr2284 tll0623 tlr0846 tsr0534 tll2275 tlr1134 tlr1258 tlr0784 tlr0417 tlr0249 tll0231 tll1029 tll1641 tlr1996 tll1495 tlr1765 tll0747 tll1092 tll0578 tlr0741 tlr0742 tll0233 tlr0707 tlr0603 tll0559 tll0180 114.2139 121.6082 113.1949 122.7854 101.164 111.1482 120.4957 122.6973 121.5234 122.6257 119.4415 116.2789 118.3729 118.3731 120.4595 120.4598 122.7491 111.1627 111.1628 119.4368 119.4297 109.1074 100.14 101.162 103.311 118.3785 120.4498 119.4464 105.572 122.7780 121.5778 122.6917 116.3032 121.6205 118.3884 120.4700 115.2456 111.1595 111.1597 121.6192 121.5526 97.8646 112.1856 118.3572 1227 1240 1246 1248 1254 1255 1256 1261 1262 1263 1265 1267 1274 1275 1278 1279 1280 1288 1289 1292 1294 1295 1298 1302 1308 1310 1311 1312 1316 1336 1342 1475 1480 1481 1485 1489 1494 1495 1496 1505 1520 1536 1541 1542 232.1183 232.5030 232.3164 232.938 232.2005 232.3779 232.2602 232.4445 232.2416 232.2415 232.638 232.4135 232.4848 232.4850 232.4042 232.4043 232.4370 232.2278 232.2277 232.3394 232.1368 232.2024 232.677 232.3437 232.3177 232.3254 232.2116 232.4363 232.962 232.3186 232.712 232.1917 232.4800 232.1729 232.2135 232.3919 232.3475 232.768 232.769 232.3763 232.1052 232.3906 232.3211 232.3215 350.3932 362.6634 334.3374 345.3716 362.6328 342.3636 354.4146 360.4940 361.5589 355.4172 361.5857 311.2740 359.4700 359.4701 344.3711 353.4096 361.5651 357.4426 235.1397 362.6606 360.5148 300.2505 362.6560 360.4897 342.3651 319.2950 360.5174 202.1000 361.5797 307.2653 329.3216 361.5475 324.3086 359.4740 353.4121 258.1727 180.783 358.4517 261.1772 266.1852 147.506 245.1540 359.4743 361.5365 pheT cobN uvrC coaD dapF leuS pgi purN argC ribA petF2 htpG rpl28 dxs pykF purK aroB nadA trpS thrB folB glgB hemE hisIE dnaA Phenylalanyl-tRNA synthetase beta subunit cobalamin biosynthetic protein CobN Excinuclease ABC subunit C (UvrC) putative pantetheine-phosphate adenylyltransferas diaminopimelate epimerase leucyl-tRNA synthetase glucose-6-phosphate isomerase phosphoribosylglycinamide formyltransferase N-acetyl-gamma-glutamyl-phosphate reductase possible GTP cyclohydrolase II probable methylthioadenosine phosphorylase conserved hypothetical protein Ferredoxin Inositol monophosphatase family protein heat shock protein HtpG 50S ribosomal protein L28 Alkyl hydroperoxide reductase conserved hypothetical protein conserved hypothetical protein 1-deoxy-D-xylulose 5-phosphate synthase conserved hypothetical protein conserved hypothetical membrane protein pyruvate kinase conserved hypothetical protein phosphoribosylaminoimidazole carboxylase 3-dehydroquinate synthase conserved hypothetical protein conserved hypothetical protein Quinolinate synthetase A protein conserved hypothetical protein putative endonuclease t-RNA synthetase, class Ib:Tryptophanyl-tRNA sy Putative glucokinase putative homoserine kinase possible dihydroneopterin aldolase conserved hypothetical protein 1,4-alpha-glucan branching enzyme Uroporphyrinogen decarboxylase (URO-D) conserved hypothetical protein histidine biosynthesis bifunctional protein HisIE conserved hypothetical protein chromosomal replication initiator protein DnaA putative multidrug efflux ABC transporter possible ATP phosphoribosyltransferase Table 2 ort270 ort015 ort361 ort562 ort316 ort240 ort498 ort642 ort564 ort646 ort193 ort254 ort199 ort500 ort353 ort092 ort483 ort051 ort219 ort196 ort657 ort431 ort640 ort543 ort014 ort356 ort446 ort334 ort365 ort479 ort404 ort094 ort307 ort306 ort416 ort314 ort389 ort231 ort408 ort299 ort424 ort208 ort050 ort455 Pag 7.6650 4.8353 3.8031 6.4254 11.7503 4.1980 4.1614 5.1686 7.5409 7.1418 7.0170 7.8605 5.0733 4.8821 3.0155 10.8625 2.5462 4.4652 11.7647 3.6021 5.0849 3.4373 8.7114 4.5350 4.1781 8.7576 3.0337 5.8052 2.8153 5.8949 7.3519 5.3471 2.7912 2.3021 2.6303 9.2178 3.1462 7.5093 3.5785 2.8794 2.1290 1.8307 7.7359 5.8213 0 0 1 0 0 1 1 1 0 0 0 0 0 1 1 0 1 0 0 1 1 1 0 1 1 0 1 0 1 0 0 1 0 1 1 0 1 0 1 1 1 1 0 0 all0580 alr4907 all4936 all0082 all3722 all4751 all4845 alr4846 alr4848 alr1343 asr1344 alr3885 alr0912 alr1956 alr1952 all2707 alr2708 all1359 all0859 alr0577 alr4746 alr4745 alr0175 all0174 alr1080 alr1026 alr1312 all0980 alr3858 all1522 all4956 alr4957 all4358 all4357 all2315 alr2082 all1985 alr5252 alr2895 all4480 all2508 asr1309 alr3210 all1663 gll3339 gll3101 gll3141 gll1250 gll0378 gll3525 glr3128 gll0444 gll0217 gll0403 gsl0402 gll1462 glr4422 gll3611 gll2639 glr3481 glr1529 gll0551 glr1731 glr0841 gll2556 glr3029 gll0554 glr3125 glr0547 glr1074 glr4015 gll0297 gll0298 gll0762 gll3085 glr3086 gll3767 gll3766 gll2657 glr2448 glr1446 gll0127 gll4314 glr1809 glr2763 gsl2877 gll3950 gll3492 Online Supporting Information 0559 1263 1264 1265 1268 1269 1270 1271 1273 1274 1275 1278 1281 1284 1286 1287 1288 1291 1292 1293 1297 1298 1299 1300 1301 1302 1305 1306 1309 1310 1311 1312 1313 1314 1315 1316 1165 0431 0430 0428 0420 0418 0415 0413 1194 0379 0378 0375 0370 0368 0367 0366 0364 0363 0362 0358 0355 0351 0349 0348 0347 0344 0343 0341 0336 0335 0334 0332 0331 0330 0323 0322 0319 0318 0317 0316 0315 0314 0313 0312 0283 0276 0274 0272 0257 0254 0233 0230 497.164 475.90 391.13 415.14 490.103 505.119 427.15 427.33 427.36 507.192 507.193 432.28 506.199 420.3 497.173 493.84 493.68 458.2 424.30 497.24 505.106 505.105 407.13 407.15 498.138 498.56 497.129 415.32 502.61 505.93 477.85 477.33 463.67 463.68 375.2 388.7 507.46 475.52 431.41 441.50 464.47 464.31 495.2 416.11 133.1213 129.545 128.486 136.2416 135.2060 131.809 135.1880 135.1934 136.2261 134.1536 134.1537 134.1375 136.2644 135.1742 135.1738 135.2009 135.1825 133.1326 134.1371 131.881 124.107 124.106 124.105 121.9 126.240 126.241 131.850 131.851 131.854 131.856 131.868 131.857 129.594 129.595 133.1141 133.1193 125.210 134.1420 135.1890 135.1922 136.2267 136.2287 128.520 133.1169 0561 1337 1338 1339 1342 1343 1344 1345 1347 1348 1349 1352 1355 1358 1360 1361 1362 1365 1366 1367 1371 1372 1373 1374 1375 1376 1379 1380 1383 1384 1385 1386 1387 1388 1389 1390 1268 0427 0426 0424 0419 0417 0413 0412 sll1019 sll0902 sll1463 slr0066 sll0099 slr0400 sll0454 sll1108 slr1882 sll0635 ssr0102 sll1489 slr0321 slr2047 slr1531 sll0169 slr1934 sll0844 slr0819 slr1761 slr0546 slr1096 slr1673 slr0017 slr1022 sll1612 sll0996 slr1874 sll1633 slr0526 sll1917 slr1972 slr0164 slr0165 sll1363 slr1925 slr0638 sll1653 sll1893 slr0056 slr0604 ssl2667 slr2143 slr0528 tll1639 tll1106 tlr0528 tlr2152 tll0172 tll0858 tlr0690 tll1786 tll1556 tlr1947 tsl0811 tll1189 tlr0056 tll0037 tlr1973 tlr0758 tlr1169 tlr2256 tll1667 tlr1102 tll0698 tll0868 tll1968 tll0780 tlr1328 tlr0190 tlr1315 tll0523 tll2382 tlr0279 tll1192 tll0008 tll1759 tlr1071 tll2254 tlr2377 tll1034 tll2373 tlr1185 tll1539 tlr0304 tsl1293 tll2327 tlr1239 117.3507 112.1696 106.637 111.1559 107.823 95.8446 106.635 106.737 119.4305 122.6515 122.6516 122.7243 97.8579 93.8377 93.8373 97.8639 97.8597 118.3701 120.5101 103.326 95.8485 95.8483 95.8481 95.8458 122.6586 122.7876 116.2972 120.5122 117.3167 122.6229 114.2234 114.2240 121.5914 121.5915 117.3269 119.4179 120.4809 120.4913 111.1638 114.2264 98.8681 113.2079 121.6038 120.4586 1543 1586 1587 1590 1596 1599 1600 1601 1603 1604 1605 1608 1613 1616 1618 1619 1620 1622 1623 1625 1629 1630 1631 1633 1634 1635 1640 1641 1644 1645 1646 1647 1648 1649 1650 1651 1671 1674 1675 1677 1684 1687 1689 1690 232.4218 232.1162 232.1196 232.5308 232.2457 232.895 232.1086 232.1087 232.1089 232.3884 232.3883 232.787 232.3381 232.3223 232.3226 232.3144 232.3145 232.2942 232.3328 232.4221 232.900 232.901 232.5289 232.5288 232.2623 232.2579 232.1955 232.4248 232.815 232.2794 232.1185 232.1184 232.5162 232.5161 232.4034 232.2041 232.3205 232.5275 232.4874 232.2245 232.4326 232.1952 232.3718 232.5010 357.4363 357.4406 251.1628 233.1372 258.1729 69.89 305.2609 300.2497 362.6378 346.3783 346.3784 284.2156 321.3013 357.4455 261.1775 226.1278 226.1283 351.4024 354.4153 282.2138 362.6707 300.2494 185.829 185.821 360.5161 355.4176 351.3979 357.4357 356.4290 318.2924 361.5610 361.5635 360.4873 360.4874 359.4730 279.2057 262.1794 359.4679 253.1658 362.6019 157.582 360.5037 316.2875 346.3769 argF ftsH3 ribD cobL pheS surE ribF thiE phoH ffh pdhA trpC lpdA spoU murA argD folC miaB ddlB ftsZ panB hemN clpP4 clpP3 ilvC cobD glyQ hisF chlG lepA murE Putative hydroxyacylglutathione hydrolase ornithine carbamoyltransferase cell division protein FtsH3 riboflavin biosynthesis protein putative precorrin-6y methylase predicted sugar kinase Phenylalanyl-tRNA synthetase:Aminoacyl tRNA s Survival protein SurE putative riboflavin kinase/FAD synthase Thiamine monophosphate synthase (TMP) DUF170 putative circadian phase modifier CpmA homolog GTP-binding protein ERA homolog PhoH-like phosphate starvation-inducible protein signal recognition particle protein (SRP54) conserved hypothetical protein Pyruvate dehydrogenase E1 alpha subunit tRNA (5-methylaminomethyl-2-thiouridylate)-methyl possible apolipoprotein n-acyltransferase FKBP-type peptidyl-prolyl cis-trans isomerase (PP Indole-3-glycerol phosphate synthase:Proteins bin putative dihydrolipoamide dehydrogenase tRNA/rRNA methyltransferase (SpoU) UDP-N-glucosamine 1-carboxyvinyltransferase putative N-acetylornithine aminotransferase putuative bifunctional Dihydrofolate/Folylpolyglutam 2-methylthioadenine synthetase D-alanine--D-alanine ligase cell division protein FtsZ putative Ketopantoate hydroxymethyltransferase possible oxygen independent coproporphyrinogen III conserved hypothetical protein ATP-dependent Clp protease proteolytic subunit 4 ATP-dependent Clp protease proteolytic subunit 3 ketol-acid reductoisomerase putative cobalamin biosynthetic protein glycyl-tRNA synthetase alpha subunit possible menaquinone biosynthesis methyltransferase Imidazole glycerol phosphate synthase subunit His chlorophyll synthase 33 kD subunit GTP-binding protein LepA NifU-like protein putative L-cysteine/cystine lyase UDP-N-acetylmuramyl-tripeptide synthetase Table 2 Pag ort095 ort621 ort228 ort312 ort096 4.0832 2.7493 8.3158 5.5048 8.7226 1 0 0 0 0 asr1611 alr2737 all3075 alr2377 asr2378 gsl1913 gll3572 glr2715 glr1772 gsr1647 0411 0410 1159 1160 1161 0228 0227 1181 1183 1184 483.85 468.69 493.24 506.211 506.212 133.1167 133.1312 136.2413 136.2673 135.1904 0410 0409 1257 1259 1260 ssl0331 slr0469 sll0905 slr0618 ssr1041 tsr0564 tlr0147 tlr1302 tll1716 tsl1557 122.6916 116.2921 121.6055 120.4741 120.4743 1693 1694 1702 1704 1705 232.3092 232.3172 232.4708 232.4094 232.4095 357.4321 203.1009 289.2247 284.2173 284.2174 ort555 7.9203 0 alr1526 glr2158 0551 1204 454.3 133.1285 0552 slr0012 tll1504 117.3212 1717 232.2790 310.2716 ort553 ort097 ort303 ort297 ort302 1.8375 4.1880 8.4178 8.7604 4.5780 1 1 0 0 0 alr1524 all3257 all5076 alr3441 all5078 glr2156 glr1519 gll2369 glr0215 gll2370 0550 0547 0545 0544 0543 1205 1208 1215 1216 1217 454.1 464.26 431.26 498.157 431.23 133.1284 136.2832 133.1278 135.2069 133.1277 0551 0548 0546 0545 0544 slr0009 slr0169 slr0750 slr0772 slr0749 tll1506 tlr0311 tll2345 tll2392 tll2347 117.3210 113.1896 122.6336 117.3130 122.6334 1718 1721 1723 1724 1725 232.2792 232.3757 232.1291 232.2368 232.1293 310.2714 195.926 351.4018 199.967 351.4016 ort481 ort060 ort659 ort001 ort098 ort099 ort100 ort415 ort401 ort319 ort624 ort643 ort362 ort230 ort313 ort258 ort505 ort515 ort532 ort012 ort511 ort636 ort399 ort052 ort101 ort102 ort194 ort403 ort103 ort104 ort441 4.3930 7.1721 7.9147 3.4536 3.9756 3.6335 9.4567 3.4278 3.7613 4.3761 4.2273 3.8232 7.5192 11.9224 5.8107 9.0805 3.6335 5.5852 2.8405 3.6693 6.2271 2.8669 3.9627 9.8892 3.3443 7.6973 6.4252 1.9856 3.3323 5.8508 3.0569 1 0 0 1 1 1 0 1 1 1 1 1 0 0 0 0 1 0 1 1 0 1 1 0 1 0 0 1 1 0 1 all1743 alr1961 all5288 alr5285 alr5284 alr5283 all5255 all4613 alr3751 all3802 alr1208 all2563 alr0718 all3785 alr3338 alr3339 all3570 alr5053 alr4784 alr1245 all4248 all5263 alr1878 asr4549 alr1093 all0200 all5306 alr3265 all4664 all4665 all1019 glr2486 glr3295 gll0754 gll2864 gll3145 gll3146 gll3765 glr3279 gll0839 glr0385 glr0288 gll1597 glr2929 gll3175 gll0771 gll3822 glr4227 gll2525 glr3280 gll3931 gll0025 glr2572 glr3212 gsl1645 glr3817 gll0542 glr0140 glr0071 gll2901 glr0255 glr0412 0542 0539 0537 0534 0533 0532 0528 0526 0525 0523 0521 0519 0518 0516 0515 0514 0511 0508 0506 0499 0497 0496 0495 0491 0490 0487 0484 0483 0480 0479 0477 1218 1224 1226 1229 1230 1231 1235 1239 1240 1243 1245 1248 1249 1251 1252 1253 1257 1259 1261 1268 1271 1272 1273 1283 1284 1291 1294 1296 1301 1302 1305 474.17 483.31 468.14 468.73 468.72 468.71 455.47 468.52 397.2 505.29 450.14 424.11 459.5 506.47 489.101 489.102 508.104 418.28 487.84 506.18 509.85 483.42 402.15 373.14 452.27 472.89 474.61 505.173 464.2 435.48 498.108 129.613 133.1209 136.2785 135.1627 135.1626 135.1625 136.2749 136.2635 136.2632 126.245 136.2827 131.907 136.2266 136.2264 136.2818 136.2819 133.1224 135.1834 125.177 133.1164 127.380 127.325 126.250 135.2158 126.244 131.839 136.2536 127.376 132.1041 132.1040 125.149 0543 0540 0538 0534 0533 0532 0528 0526 0525 0523 0521 0519 0518 0516 0515 0514 0510 0508 0506 0499 0497 0495 0494 0490 0489 0486 0483 0482 0478 0477 0475 slr0506 sll0809 sll0356 sll0728 sll0209 sll0208 slr1573 slr2088 slr0839 slr0260 sll0145 slr1793 sll1833 slr0959 slr1124 sll1018 slr1622 sll1425 sll1823 slr1898 sll0270 slr0653 slr1887 ssl2296 slr0609 slr1411 sll1854 sll0017 slr1926 slr0039 sll0555 tlr0575 tlr0228 tll2272 tll1311 tll1312 tll1313 tll2307 tlr1296 tlr2216 tlr0265 tll0259 tll0467 tlr2074 tlr1890 tlr1532 tlr1533 tll1883 tlr0777 tll0531 tll1408 tll0415 tll0617 tll1646 tsl1825 tlr2276 tll2296 tlr1785 tlr0479 tlr0863 tlr2026 tll0424 122.7943 122.7442 120.4765 119.4335 122.6742 122.6741 113.2045 121.5292 117.3282 116.2714 117.3155 121.5841 115.2584 109.1219 109.1081 109.1083 122.7810 112.1826 121.5788 101.151 104.436 97.8595 122.6982 91.8284 122.7669 118.3643 94.8387 121.5195 121.6188 121.6187 121.5861 1726 1731 1733 1736 1737 1738 1742 1746 1747 1750 1753 1759 1760 1762 1763 1764 1769 1771 1773 1780 1782 1783 1785 1794 1795 1802 1807 1809 1818 1819 1822 232.4185 232.3219 232.1491 232.1488 232.1487 232.1486 232.5272 232.1693 232.5423 232.873 232.4506 232.4377 232.3478 232.5395 232.2549 232.2548 232.2448 232.1272 232.1024 232.4435 232.5055 232.5263 232.3629 232.1464 232.2611 232.1631 232.1511 232.3765 232.970 232.969 232.2573 318.2919 358.4539 359.4710 235.1404 334.3385 361.5731 184.818 361.5428 291.2281 289.2248 113.282 346.3782 203.1021 335.3409 335.3401 326.3134 288.2235 357.4342 357.4351 362.6710 321.3016 361.5683 295.2369 362.6672 361.5580 362.6650 262.1783 307.2647 162.628 361.5799 358.4501 Online Supporting Information rpsD cobB cbbS, rbcS cbbL, rbcL chlN chlB chlL pcr, por trpF accA ilvB hemH cobO rrf tal ftsI cobC ppa proS purA argB priA sigA hemC hemL map conserved hypothetical protein 30S ribosomal protein S4 possible Maf-like protein implicated in septum forma Cobyric acid synthase CobB conserved hypothetical protein Ribulose bisphosphate carboxylase, small chain Ribulose bisphosphate carboxylase, large chain conserved hypothetical protein possible light-independent protochlorophyllide reduct light-independent protochlorophyllide reductase ChlB Protochlorophyllide reductase iron-sulfur Light dependent protochlorophyllide oxido-reduct Biotin/lipoate A/B protein ligase family phosphoribosylanthranilate isomerase acetyl-CoA carboxylase, alpha subunit conserved hypothetical protein conserved hypothetical protein conserved hypothetical protein acetolactate synthase Ferrochelatase possible cob(I)alamin adenosyltransferase ribosome recycling factor protein Rrf transaldolase putative peptidoglycan synthetase (pbp transpeptidase Possible membrane associated protease putative alpha-ribazole-5'-P phosphatase putative dihydroorotase inorganic pyrophosphatase Prolyl-tRNA synthetase Adenylosuccinate synthetase acetylglutamate kinase primosomal protein N' (replication factor Y) Putative principal RNA polymerase sigma factor Porphobilinogen deaminase 4a-hydroxytetrahydrobiopterin dehydratase (PCD) conserved hypothetical protein conserved hypothetical protein exodeoxyribonuclease III glutamate-1-semialdehyde 2,1-aminomutase conserved hypothetical protein conserved hypothetical protein putative methionine aminopeptidase Table 2 ort583 ort387 ort289 ort250 ort488 ort486 ort680 ort317 ort508 ort670 ort421 ort350 ort277 ort275 ort325 ort105 ort563 ort279 ort106 ort653 ort467 ort040 ort668 ort107 ort271 ort108 ort635 ort610 ort503 ort284 ort198 ort459 ort023 ort520 ort360 ort489 ort487 ort447 ort448 ort449 ort109 ort402 ort521 ort615 Pag 2.4088 5.2403 8.4611 4.4184 3.0326 3.9123 5.2465 5.2407 5.6955 8.3222 7.0685 3.6287 4.6512 4.8385 8.3212 9.5613 6.6652 7.2745 8.5202 4.6648 3.4549 10.9163 6.4108 4.4849 4.0526 4.9635 4.2058 2.4249 3.1232 5.9013 4.5751 6.6462 5.8973 3.2338 5.8556 1.7277 1.1236 ###### 3.9291 4.1129 9.1011 9.2934 2.1642 1.6101 1 0 0 1 0 0 0 0 0 0 0 1 1 0 0 0 1 0 0 0 1 0 0 0 0 0 1 1 1 0 1 0 0 1 0 1 1 0 1 1 0 0 1 1 alr5297 all3205 all4374 all2456 all2453 all2452 all4699 all4698 all3552 alr3553 all5167 alr1894 alr2373 alr2372 all0948 all0949 all5258 alr5260 alr0297 alr2780 all4883 alr5256 all4012 alr3414 alr3415 all4296 all3976 all3969 all4396 all4680 alr3739 all0049 alr2782 asr4319 alr4320 alr3422 alr3421 alr3455 alr3456 asr3457 alr0116 alr0115 all0138 all0136 glr0828 gll2378 glr1622 gll1912 glr3038 glr3039 gll4393 gll1377 gll0510 glr3428 glr2791 glr3506 glr2168 glr2167 glr2166 glr2165 glr3604 gll3752 glr4139 glr3198 glr3120 gll1338 glr2705 glr1476 glr2231 glr1649 glr0716 gsl3914 gll3149 gll3021 glr4375 glr3637 glr2442 gsl3408 gll3407 gll1918 gll1919 glr1992 glr1993 gsr1994 glr3274 glr1755 glr2999 glr0987 Online Supporting Information 0475 0473 0466 0464 0462 0461 0460 0459 0457 0456 0454 0453 0450 0449 0448 0447 0442 0441 0439 0436 0435 0433 1181 1185 1186 1187 1188 1190 1191 1193 0218 0217 0387 0329 0328 0326 0325 0322 0321 0320 0319 0318 0315 0312 1306 1308 1319 1320 1322 1323 1324 1325 1327 1328 1330 1333 1337 1338 1339 1340 1346 1348 1350 1353 1354 1359 1366 1370 1371 1373 1374 1376 1377 1379 1544 1546 0195 0187 0186 1648 1649 1652 1653 1654 1659 1660 1665 1668 354.22 345.1 482.129 506.80 506.86 506.87 452.58 452.59 482.64 482.59 498.69 362.19 506.208 506.206 493.120 493.119 455.45 455.15 484.95 482.133 456.29 455.12 412.23 507.95 507.96 507.100 357.10 357.13 375.11 422.12 398.35 506.160 373.1 452.35 452.37 509.270 509.269 504.122 504.123 504.124 370.3 370.2 494.8 494.9 129.556 131.864 134.1450 135.2188 124.82 124.83 124.84 124.85 135.1837 135.1999 127.311 127.402 130.661 130.660 136.2540 136.2539 135.2166 134.1436 131.927 133.1274 133.1206 136.2348 135.2022 135.1642 135.1643 135.1646 136.2288 136.2291 125.155 136.2657 126.261 127.354 126.288 128.451 128.452 131.827 131.826 135.1975 126.213 126.214 135.1790 135.1789 127.302 127.306 0473 0471 0464 0462 0460 0459 0458 0457 0456 0455 0453 0452 0446 0445 0444 0443 0438 0437 0435 0432 0431 0429 1297 1301 1302 1303 1304 1306 1307 1309 0246 0245 0384 0371 0370 0368 0367 0364 0363 0362 0360 0359 0354 0351 sll1740 sll0179 slr0880 sll0194 sll1316 sll1317 sll1187 slr0239 sll1546 slr0926 slr0951 slr0886 sll0760 sll0759 sll1899 sll1898 sll0300 slr1520 sll1500 slr2058 sll0223 sll1655 sll0109 slr1470 slr1471 slr0480 slr1703 slr0628 sll1043 sll0818 slr1090 sll1772 sll1112 ssr2831 slr1689 slr0343 slr0342 sll0288 sll0289 ssl0546 sll1866 sll1237 slr0906 slr1356 tlr1303 tll0506 tll2058 tlr0953 tlr0959 tlr0960 tll2157 tlr0563 tll1680 tlr0402 tlr0605 tlr1502 tll1324 tlr0010 tll1893 tll1894 tlr1686 tlr2042 tll2306 tll1881 tll0045 tll1111 tll1803 tll0601 tll0600 tll0076 tlr1552 tll1826 tll2193 tlr1514 tll2353 tlr2472 tlr0494 tsl1567 tll1566 tlr0797 tlr0796 tlr2016 tlr2017 tlr2018 tll2123 tlr1836 tlr1530 tll2046 121.5555 121.5357 121.5283 115.2553 115.2492 115.2493 122.6320 122.6321 113.1977 113.1993 118.3942 121.6008 108.944 120.5080 115.2523 115.2522 122.6503 122.7629 122.6319 122.7660 93.8370 113.1937 105.523 122.7227 122.7228 122.7231 106.673 121.5779 122.6329 122.7874 120.5042 121.5587 122.6607 121.6202 121.6203 101.117 101.116 113.2066 113.2067 113.2068 116.2865 116.2862 122.6941 122.6943 1824 1826 1838 1839 1841 1842 1843 1844 1846 1847 1849 1852 1857 1858 1859 1860 1865 1867 1869 1872 1873 1876 1885 1889 1890 1892 1894 1896 1897 1899 1928 1931 1938 1960 1961 1966 1967 1970 1971 1972 1975 1976 1982 1985 232.1502 232.3713 232.2209 232.4275 232.4273 232.4272 232.939 232.940 232.2430 232.2431 232.1369 232.2657 232.4090 232.4089 232.4409 232.4410 232.5269 232.5267 232.1981 232.3932 232.1122 232.5271 232.673 232.2337 232.2338 232.5105 232.705 232.713 232.2185 232.959 232.5435 232.1592 232.3934 232.5127 232.5128 232.2345 232.2344 232.2379 232.2380 232.2381 232.5335 232.5334 232.5357 232.5355 289.2249 361.5986 360.4952 357.4361 201.991 319.2936 360.5189 327.3183 272.1948 361.5393 289.2244 292.2303 362.6704 362.6673 362.6661 362.6660 306.2643 306.2627 360.5199 358.4636 362.6309 361.5316 268.1885 359.4795 359.4796 252.1632 257.1711 263.1798 358.4563 204.1029 171.702 331.3287 361.5527 118.312 118.313 356.4240 356.4239 155.566 155.567 155.568 247.1572 358.4534 77.123 335.3414 rpl19 gltX petC petA yvoC cobM ppx ubiA ispD fabG ctaB ribE topA ndhB birA tyrA serRS rps14 pnp mutS aroD psaE fpg petD petB minC minD minE hemK psbB rps1a possible 50S ribosomal protein L19 glutamyl-tRNA synthetase similar to secreted protein MPB70 precursor protein secretion component, Tat family Cytochrome b6f complex subunit (Rieske iron-sulfur p apocytochrome f putative prolipoprotein diacylglyceryl transferase PUTATIVE PRECORRIN-4 C11-METHYLTRANSF putative exopolyphosphatase probable 4-hydroxybenzoate-octaprenyltransferase putative 2-C-methyl-D-erythritol 4-phosphate cytidyl 3-oxoacyl-[acyl-carrier protein] reductase putative multidrug efflux ABC transporter putative multidrug efflux ABC transporter putative protoheme IX farnesyltransferase conserved hypothetical protein Putative Riboflavin synthase alpha chain putative oxidoreductase, aldo/keto reductase family conserved hypothetical protein Prokaryotic DNA topoisomerase NADH dehydrogenase I chain 2 (or N) putative Biotin--acetyl-CoA-carboxylase ligase Chorismate mutase conserved hypothetical protein Putative inner membrane protein conserved hypothetical protein putative seryl-tRNA synthetase 30S ribosomal protein S14 polyribonucleotide nucleotidyltransferase putative tetrapyrrole methylase family protein GTP1/Obg family GTP-binding protein putative DNA mismatch repair protein MutS family Dehydroquinase class II photosystem I subunit IV (PsaE) Formamidopyrimidine-DNA glycolase (FAPY-DNA g cytochrome b6f complex subunit 4 (17 kd polypeptid apocytochrome b6 possible septum site-determining protein putative septum site-determining protein MinD possible septum site-determining protein MinE conserved hypothetical protein possible protoporphyrinogen oxidase photosystem II chlorophyll-binding protein CP47 30S ribosomal protein S1, homolog A Table 2 ort444 ort252 ort274 ort480 ort540 ort568 ort110 ort206 ort111 ort519 ort658 ort112 ort506 ort557 ort113 ort450 ort114 ort212 ort241 ort115 ort116 ort556 ort117 ort272 ort118 ort593 ort597 ort588 ort584 ort614 ort587 ort617 ort580 ort592 ort612 ort601 ort589 ort598 ort620 ort599 ort582 ort622 ort579 ort633 Pag 2.6998 7.5240 6.7345 6.4817 11.4171 5.2948 9.6354 4.7836 8.9527 4.1518 4.6495 6.3363 4.1691 6.4191 5.9256 2.7706 7.4787 6.2936 9.1785 9.9210 7.5851 1.8121 11.0019 7.5610 5.7911 3.4338 4.2138 4.8482 2.8292 1.6236 2.7470 2.3864 1.7309 5.4025 3.2755 2.2454 3.6335 2.0908 3.2022 4.2082 4.0830 2.3137 4.0651 3.3749 1 0 0 0 0 0 0 1 0 1 0 0 1 0 0 1 0 0 0 0 0 1 0 0 0 1 1 1 1 1 1 1 1 0 1 1 1 1 1 1 1 1 1 1 alr4124 all4656 alr5068 alr2936 alr3525 all3791 alr3980 alr0652 alr2572 all0329 all0328 alr3351 all4861 all3374 alr1286 all2906 alr5126 all5123 all1141 alr4075 asr4076 all3272 alr3863 alr2308 alr4216 all4215 all4214 all4213 all4212 asl4211 all4210 all4209 all4208 asl4207 asl4206 all4205 asl4204 all4203 all4202 all4201 all4200 all4199 all4198 all4197 gll2577 gll0781 gll3174 gll1806 gll0720 gll3123 glr3763 glr1480 glr1586 glr3701 glr1717 glr2275 gll0414 gll2405 gll2047 gll3412 gll3173 gll2874 glr0406 glr1449 gsl3135 gll3416 glr2043 gll3769 gll3770 glr0085 glr0086 glr0087 glr0903 gsr0904 gll3922 gll3921 gll3920 gsl3919 gsl3918 gll3917 gll3916 gll3915 gll3913 gll3912 gll3911 gll3910 gll3909 gll1393 Online Supporting Information 0311 0310 1591 1590 1589 1584 1583 1581 1579 1578 1577 1576 1575 1573 1571 1569 1567 1566 1565 1564 1563 1562 1561 1560 1559 1558 1557 1556 1555 1554 1553 1552 1551 1550 1549 1548 1547 1546 1545 1544 1543 1542 1541 1540 1670 1671 1688 1689 1691 1703 1704 1707 1709 1710 1711 1712 1713 1715 1717 1719 1722 1723 1724 1725 1726 1727 1728 1730 1731 1732 1733 1734 1735 1736 1737 1738 1739 1740 1741 1742 1743 1744 1745 1746 1747 1748 1749 1750 474.106 464.9 418.37 461.69 461.72 505.114 357.7 302.2 421.30 485.8 485.9 346.3 458.16 450.37 456.38 391.20 386.17 406.9 389.1 507.14 507.16 324.3 372.20 481.68 374.3 374.12 374.13 374.14 374.15 374.16 374.17 374.18 374.19 374.20 374.21 374.22 374.23 374.24 374.25 374.27 374.28 374.29 374.30 374.31 129.637 129.638 136.2278 134.1385 132.1028 127.397 127.313 134.1416 133.1150 134.1500 134.1501 131.939 131.940 131.942 133.1260 136.2440 135.1986 126.252 127.383 127.317 127.318 136.2347 127.367 127.363 130.685 130.702 130.703 130.704 130.705 130.706 130.707 130.708 130.709 130.710 130.711 130.712 130.713 130.714 130.715 130.716 130.717 130.718 130.719 130.720 0349 0348 1747 1746 1745 1740 1739 1736 1734 1733 1732 1731 1730 1728 1726 1724 1721 1720 1719 1718 1717 1716 1715 1714 1713 1712 1711 1710 1709 1708 1707 1706 1705 1704 1703 1702 1701 1700 1699 1698 1697 1696 1695 1694 sll0927 slr1420 slr0328 sll1249 slr0838 sll0320 slr0589 slr0067 slr1285 slr0737 slr0738 slr0990 sll0920 sll1277 slr1717 sll1536 slr1563 slr1293 slr2081 slr1495 ssr0332 sll0569 sll0295 sll0031 sll1262 sll1799 sll1800 sll1801 sll1802 ssl3432 sll1803 sll1804 sll1805 ssl3436 ssl3437 sll1806 sll1807 sll1808 sll1809 sll1810 sll1811 sll1812 sll1813 sll1814 tll0978 tll1291 tlr1810 tll2450 tlr1811 tlr1083 tll0625 tlr2419 tlr1565 tll1724 tll1672 tll0322 tll1912 tll2444 tll1241 tlr2403 tlr1821 tll0232 tll2435 tlr1240 tsl2097 tlr2089 tlr1691 tlr0739 tlr1130 tlr0081 tlr0082 tlr0083 tlr0084 tsr0085 tlr0086 tlr0087 tlr0088 tsr0089 tsr0090 tlr0091 tlr0092 tlr0093 tlr0094 tlr0095 tlr0096 tlr0097 tlr0098 tlr0099 122.6945 119.4142 120.4679 121.6145 121.5252 116.2883 120.4770 101.90 119.4342 117.3514 117.3515 121.5582 116.3099 111.1601 122.7215 108.980 99.8746 117.3387 122.6634 122.7496 122.7498 122.6631 122.6543 99.8758 99.8757 99.8766 99.8767 99.8768 99.8769 99.8770 99.8771 99.8772 99.8773 99.8774 99.8775 99.8776 99.8777 99.8778 99.8779 99.8780 99.8781 99.8782 99.8783 99.8784 1987 1988 2026 2027 2029 2036 2038 2041 2043 2044 2045 2046 2047 2049 2051 2053 2056 2057 2058 2059 2060 2062 2063 2065 2066 2067 2068 2069 2070 2071 2072 2073 2074 2075 2076 2077 2078 2079 2080 2081 2082 2083 2084 2085 232.4655 232.977 232.1284 232.4831 232.2123 232.889 232.702 232.3443 232.4385 232.3578 232.3577 232.2104 232.1104 232.2294 232.1930 232.4861 232.1299 232.1302 232.3264 232.2057 232.2056 232.3769 232.811 232.4026 232.4566 232.4567 232.4568 232.4569 232.4570 232.4571 232.4572 232.4573 232.4574 232.4575 232.4576 232.4577 232.4578 232.4579 232.4580 232.4581 232.4582 232.4583 232.4584 232.4585 361.5852 255.1690 231.1345 362.6150 312.2767 360.5036 321.3009 205.1043 358.4523 344.3693 344.3694 315.2858 297.2431 269.1897 326.3155 151.536 332.3320 362.6111 357.4419 225.1269 361.5834 76.118 360.5108 339.3546 362.6376 298.2439 298.2440 298.2441 298.2442 298.2443 298.2444 298.2445 298.2446 298.2447 298.2448 298.2449 296.2389 296.2390 296.2391 296.2392 296.2393 296.2394 296.2395 296.2396 metK panC purM rnd psaD trpE ppc recF moeB recA rpl3 rpl4 rpl23 rpl2 rps19 rpl22 rps3 rpl16 rpl29 rps17 rplN rpl24 rpl5 rps8 rpl6 rpl18 rpsE rpl15 secY S-adenosylmethionine synthetase Putative carbohydrate kinase, FGGY family putative low molecular weight protein-tyrosine-phosp putative bifunctional enzyme; pantothenate synthetase phosphoribosyl formylglycinamidine cyclo-ligase putative ribonuclease D conserved hypothetical protein MRP protein homolog conserved hypothetical protein photosystem I reaction center subunit II (PsaD) Anthranilate synthase component I and chorismate bin conserved hypothetical protein phosphoenolpyruvate carboxylase putative DNA repair and genetic recombination prote conserved hypothetical protein molybdopterin biosynthesis protein conserved hypothetical protein phytoene dehydrogenase related enzyme Prephenate dehydrogenase conserved hypothetical protein conserved hypothetical protein RecA bacterial DNA recombination protein conserved hypothetical protein putative ldpA protein conserved hypothetical protein 50S ribosomal protein L3 50S ribosomal protein L4 50S ribosomal protein L23 50S ribosomal protein L2 30S ribosomal protein S19 50S ribosomal protein L22 30S ribosomal protein S3 50S ribosomal protein L16 50S ribosomal protein L29 30S ribosomal protein S17 50S ribosomal protein L14 50S ribosomal protein L24 50S ribosomal protein L5 30S ribosomal protein S8 50S ribosomal protein L6 50S ribosomal protein L18 30S ribosomal protein S5 50S ribosomal protein L15 preprotein translocase SecY subunit Table 2 ort007 ort609 ort607 ort602 ort581 ort661 ort578 ort202 ort011 ort517 ort516 ort047 ort429 ort385 ort608 ort619 ort368 ort665 ort606 ort059 ort497 ort570 ort569 ort119 ort328 ort278 ort237 ort281 ort222 ort120 ort462 ort121 ort223 ort122 ort123 ort025 ort236 ort125 ort126 ort127 ort128 ort571 ort412 ort291 Pag 6.9091 2.3989 1.2622 3.7192 2.1533 6.1436 2.9475 2.8099 6.0994 1.3910 2.2075 9.9441 3.3616 3.8422 0.9678 2.8512 1.9792 1.7116 0.8543 4.7356 6.1800 7.7365 3.1798 3.9344 9.5744 7.9381 7.8222 7.6511 5.1723 9.7074 6.8213 8.4493 3.5794 6.3493 5.8153 6.5786 5.3009 5.2226 4.0985 7.1284 4.8971 2.0237 6.2940 9.3496 0 1 1 1 1 0 1 1 0 1 1 0 1 1 1 1 1 1 1 1 1 0 1 0 0 0 0 1 0 0 0 0 1 0 0 0 0 0 1 0 1 1 0 0 all4196 all4193 all4192 all4191 all4190 all4189 all4188 all2457 alr2458 alr5155 alr5154 all0453 all1694 alr4344 all4340 all4339 all4338 all4337 all4336 all4335 alr4334 alr4332 alr4331 asl4325 all4464 alr4094 all1802 all1800 all1211 alr1215 alr1217 all1220 alr1222 all4101 all4102 alr1244 alr0063 all2381 alr2385 alr2386 asr4321 all0129 all4705 all4708 gll1392 gll3574 gll3573 gll3571 gll3570 gll3569 glr4418 glr3730 glr3818 glr3439 glr3438 gll2546 gll1029 glr1508 glr3925 glr3926 glr3927 glr3928 glr3929 glr3968 gll0524 glr1507 glr1506 gsl0746 glr1656 glr3503 glr0515 glr0516 glr2775 glr3755 glr0846 glr2112 gll4317 gll0853 gll4038 glr2710 glr4032 gll0842 gll4382 glr3390 gsl3544 gll4162 gll1432 glr0678 Online Supporting Information 1539 1537 1536 1535 1534 1533 1532 1528 1527 1523 1524 1525 1514 1512 1511 1510 1509 1508 1507 1506 1504 1502 1501 1499 0081 0082 0084 0085 0098 0099 0100 0101 0102 0103 0104 0107 0112 0114 0116 0117 0121 0128 0129 0130 1751 1753 1754 1755 1756 1757 1758 1762 1763 1769 1770 1772 1775 1777 1779 1780 1781 1782 1783 1784 1786 1788 1789 1792 1624 1625 1628 1629 0181 0179 0177 0176 0175 0172 0170 0163 1963 1967 1969 1970 1975 1988 1989 1990 374.32 374.35 374.36 374.37 374.38 374.39 374.40 401.28 401.6 456.8 456.7 502.74 463.15 423.41 423.16 423.17 423.18 423.19 423.20 423.21 423.36 423.35 423.34 443.38 493.74 507.59 489.87 489.88 475.2 441.1 441.4 441.58 441.8 486.93 486.92 506.16 472.17 506.63 506.215 506.216 452.38 504.130 428.12 399.9 130.721 130.723 130.724 130.725 130.726 130.727 130.728 130.684 130.732 135.1944 135.1943 135.1783 136.2846 132.1054 132.1058 132.1060 132.1061 132.1062 132.1063 132.1064 132.1053 132.1052 132.1051 127.309 129.611 129.541 134.1554 134.1555 135.1840 133.1240 135.2046 135.1918 129.608 124.116 124.115 126.297 136.2342 133.1153 126.222 126.223 129.534 136.2478 136.2613 136.2612 1693 1691 1690 1689 1688 1687 1686 1682 1681 1673 1672 1671 1670 1668 1667 1666 1665 1664 1663 1662 1660 1658 1657 1654 0095 0096 0098 0099 0117 0118 0119 0120 0121 0123 0125 0128 0132 0135 0137 0138 0141 0150 0151 0152 sll1815 sll1816 sll1817 sll1818 sll1819 sll1820 sll1821 sll1193 slr0827 slr1835 slr1834 slr0969 slr1598 sll1499 sll1096 sll1097 sll1098 sll1099 sll1101 sll0195 sll1662 slr1130 slr1129 ssr2723 slr1791 slr1743 slr1509 sll0493 slr2030 slr1990 sll0631 slr0565 slr0082 slr0651 slr0656 sll1669 sll0754 slr0941 slr1417 sll1979 ssl1690 slr0115 slr0446 slr0379 tlr0100 tlr0103 tlr0104 tlr0105 tlr0106 tlr0107 tlr0108 tlr0496 tll0460 tlr0732 tlr0731 tll1307 tlr0613 tll1368 tlr1747 tlr1748 tlr1749 tlr1750 tlr1751 tlr2461 tlr2106 tll0663 tlr0597 tll0211 tll1035 tlr1136 tll0303 tlr0412 tll1647 tll0200 tll2408 tlr0588 tlr1564 tlr0351 tll1862 tlr0882 tll1629 tll0336 tll0867 tsl0866 tsl0017 tlr2423 tll0518 tlr2055 99.8785 99.8789 99.8790 99.8791 99.8792 99.8793 99.8794 122.6287 122.7861 122.6939 122.6938 117.3403 110.1280 122.6439 122.7700 122.7701 122.7702 122.7703 122.7704 122.7705 122.7706 122.6427 122.6426 98.8736 121.5284 119.4435 120.4792 120.4794 121.5510 120.4512 105.589 75.7991 121.5424 122.7777 122.7036 122.6361 104.471 120.4901 120.4747 120.4748 121.6204 122.6827 122.7849 122.7848 2086 2088 2089 2090 2091 2092 2093 2097 2098 2123 2124 2126 2130 2132 2135 2136 2137 2138 2139 2140 2142 2144 2145 2148 2164 2165 2168 2169 2188 2190 2192 2193 2194 2197 2198 2202 2207 2212 2214 2215 2219 2236 2237 2238 232.4586 232.4589 232.4590 232.4591 232.4592 232.4593 232.4594 232.4276 232.4277 232.1362 232.1361 232.1795 232.5025 232.5151 232.5147 232.5146 232.5145 232.5144 232.5143 232.5142 232.5141 232.5139 232.5138 232.5132 232.2234 232.4685 232.3659 232.3658 232.4509 232.4511 232.4513 232.4516 232.4518 232.4680 232.4679 232.4434 232.1604 232.4097 232.4100 232.4101 232.5129 232.5348 232.935 232.934 296.2397 296.2399 296.2400 296.2401 296.2402 296.2403 296.2404 360.5171 357.4397 221.1223 221.1222 307.2665 216.1162 361.5417 359.4692 359.4693 359.4694 359.4696 359.4697 311.2738 311.2758 362.6178 355.4186 299.2462 348.3852 362.6161 353.4116 353.4117 349.3902 310.2720 304.2580 344.3709 362.6256 362.6430 360.5112 356.4296 346.3787 361.5405 359.4851 359.4852 361.5884 362.6453 342.3628 231.1340 adk rps13 rps11 rpoA rpl17 truA rpl13 alr psaB psaA cbiG lipA glsF rps12 rps7 fusA tufA rps10 pheA rnhB rne cysH nadB aroK rpaA holB Adenylate kinase 30S ribosomal protein S13 30S ribosomal protein S11 RNA polymerase alpha subunit 50S ribosomal protein L17 tRNA pseudouridine synthase A 50S ribosomal protein L13 HNH endonuclease family protein putative alanine racemase photosystem I P700 chlorophyll a apoprotein subuni photosystem I P700 chlorophyll a apoprotein subuni bifunctional cbiH protien and precorrin-3B C17-meth lipoic acid synthetase Ferredoxin-dependent glutamate synthase, Fd-GO 30S ribosomal protein S12 30S ribosomal protein S7 elongation factor EF-G elongation factor EF-Tu 30S ribosomal protein S10 ATP-dependent protease La (LON) domain Chorismate mutase-Prephenate dehydratase ribonuclease HII ribonuclease E conserved hypothetical protein Phosphoadenosine phosphosulfate reductase putative NADH dehydrogenase, transport associated possible sodium transporter, Trk family putative potassium channel, VIC family possible Fe-S oxidoreductase conserved hypothetical protein putative L-aspartate oxidase conserved hypothetical protein possible Fe-S oxidoreductase conserved hypothetical protein conserved hypothetical protein Shikimate kinase possible Ribosome-binding factor A conserved hypothetical protein conserved hypothetical protein conserved hypothetical protein conserved hypothetical protein two-component response regulator DNA polymerase III, delta prime subunit Thymidylate kinase Table 2 Pag ort191 ort552 ort572 ort502 ort501 ort676 ort213 4.3036 3.9926 1.1578 4.3056 5.9015 6.5207 9.7292 1 0 1 1 0 0 0 all3193 alr3823 all3822 alr0238 alr0241 asr3137 all3136 glr0715 glr2406 glr2669 gll0800 gll3009 gsl4394 gll2924 0132 0133 0134 0135 0138 0140 0141 1992 1993 1994 1995 1998 2000 2001 387.10 479.20 479.118 506.153 506.157 475.30 475.75 131.820 133.1188 133.1298 133.1299 133.1302 133.1185 132.1104 0154 0155 0156 0157 0160 0163 0164 slr0823 slr0448 slr0947 slr1510 sll1848 ssr1425 sll0825 tll1525 tlr1103 tll2364 tlr0844 tlr0042 tsl2457 tlr0715 122.7024 119.4331 119.4162 115.2555 115.2561 108.1011 108.928 2240 2245 2246 2247 2250 2253 2254 232.2777 232.852 232.853 232.1666 232.1669 232.2723 232.2722 253.1646 361.5486 361.5773 358.4622 360.5035 238.1449 238.1438 ort323 ort129 ort554 ort471 ort469 ort130 ort131 ort238 ort470 ort472 ort473 ort466 ort386 ort656 ort536 ort229 ort293 ort207 ort132 ort442 ort393 ort509 ort133 ort337 ort478 ort134 ort019 ort463 ort285 ort639 ort567 ort136 4.9792 4.1746 2.8047 4.0972 2.5841 6.5891 4.9200 3.5926 1.9638 4.3225 2.6506 2.5004 4.9342 2.3165 5.4007 5.5545 4.0812 6.2389 7.1534 7.5926 7.2132 2.6452 7.6884 6.7481 4.3700 6.0225 9.8344 8.5661 5.9917 3.7358 5.3363 5.7803 0 1 1 1 1 0 0 1 1 1 1 1 0 1 1 1 1 0 0 0 1 1 0 0 1 0 0 0 0 1 1 0 alr1833 all1732 all3953 alr3956 alr3957 alr3399 all0783 asl2551 alr0226 alr0225 alr0224 alr0223 alr0222 all3794 alr0989 alr3201 all1736 alr2505 all3354 alr0033 all3859 all0273 all0271 all0270 all0269 all0268 all3717 all2091 all4677 all1549 alr0522 all0745 glr1744 glr1522 glr1340 glr0218 glr0219 glr1857 glr0789 gsl4037 gll0652 gll0653 gll0654 gll1584 glr3012 glr2758 gll1251 glr4402 glr1588 gll4384 glr2927 gll1578 gll3682 glr1233 gll1243 gll1242 glr0883 glr0884 glr4279 gll0888 glr2452 glr2748 glr0073 glr3866 0143 0145 0147 0149 0150 0151 0153 0154 0157 0158 0159 0160 0161 0164 0167 0168 0169 0170 0171 0176 0178 0180 0182 0183 0184 0185 0187 0188 0189 0191 0193 0194 2003 2005 2007 2009 2010 2011 2013 2014 2017 2018 2019 2020 2021 2027 2038 2040 2043 2045 2048 2059 2061 2063 2065 2066 2067 2068 2070 2073 2074 2111 2109 2108 474.11 434.47 429.39 429.15 429.16 506.25 492.56 454.36 401.40 401.39 401.38 401.36 401.35 505.109 372.14 507.107 434.46 461.80 498.130 382.6 502.154 480.45 480.47 480.48 480.49 480.50 507.173 372.1 501.94 405.29 456.45 482.87 135.1845 135.1987 135.1843 135.1990 135.1991 135.1994 128.445 131.930 128.471 128.470 128.469 128.468 127.342 130.657 136.2802 136.2296 135.2057 129.562 134.1484 133.1232 131.823 136.2313 136.2315 136.2316 136.2317 136.2318 135.2189 126.243 135.2184 133.1254 134.1412 134.1410 0166 0168 0170 0172 0173 0174 0176 0177 0181 0182 0183 0184 0185 0188 0191 0193 0194 0195 0196 0201 0203 0205 0207 0208 0209 0210 0212 0213 0214 0217 0219 0220 slr1255 slr1623 sll0998 slr0844 slr0331 slr1530 slr2141 ssl0564 sll0522 sll0521 sll0520 sll0519 sll0401 slr0543 sll0901 slr0525 slr1783 slr0387 sll1144 slr1518 slr1238 sll1865 slr0053 slr0054 slr0055 slr2005 sll0502 slr0936 sll1615 slr1325 slr1629 slr0267 tll1560 tll0447 tlr1206 tll0720 tll0719 tll1658 tll1771 tsl1865 tlr0670 tlr0669 tlr0668 tlr0667 tlr2393 tll2475 tll0586 tll0451 tlr1263 tlr0115 tlr2072 tlr2196 tlr0908 tll0527 tlr1585 tlr1586 tlr1471 tlr1472 tll0826 tll1713 tlr0961 tll0584 tll1538 tlr1507 120.4690 104.483 119.4263 118.3907 118.3909 101.110 122.6918 121.5189 122.6296 122.6295 122.6294 122.6291 122.6290 118.3575 121.5847 122.7900 121.5862 120.4669 120.5159 120.4641 120.4545 109.1139 118.3814 118.3815 118.3817 113.2053 120.4920 117.3252 107.835 122.7534 115.2450 113.1916 2256 2258 2260 2262 2263 2265 2267 2268 2271 2272 2273 2274 2275 2280 2285 2287 2289 2291 2298 2307 2309 2311 2313 2314 2315 2316 2319 2320 2322 2324 2326 2327 232.3649 232.4194 232.729 232.726 232.725 232.2324 232.1916 232.4368 232.1655 232.1654 232.1653 232.1652 232.1651 232.886 232.1879 232.2784 232.4192 232.4323 232.2107 232.1574 232.814 232.1720 232.1718 232.1717 232.1716 232.1715 232.2460 232.2050 232.960 232.3830 232.1850 232.3505 197.948 288.2236 294.2363 354.4160 349.3920 272.1946 361.5476 195.935 362.6240 362.6239 362.6238 362.6237 249.1600 314.2817 287.2211 318.2923 283.2148 253.1650 187.843 360.4882 360.5226 362.6601 362.6503 362.6504 362.6505 362.6506 360.5034 267.1863 351.4010 360.4975 362.6597 361.5504 ort494 ort457 ort407 ort546 2.7961 6.9532 8.1217 5.7483 0 0 0 0 all4131 alr0477 alr3936 all4272 glr2313 gll1671 gll3423 gll3080 0195 0197 0198 0199 2106 2101 2100 2094 501.49 494.62 498.63 460.17 134.1572 131.814 134.1570 136.2715 0221 0223 0224 0225 slr0394 slr1656 sll1713 slr1418 tll2268 tlr1809 tll2266 tll0579 113.2055 107.798 116.2830 119.4239 2329 2333 2334 2337 232.4647 232.1819 232.746 232.5080 361.5524 361.5832 361.5256 356.4298 Online Supporting Information ycf3 radA rpaB plsX plsC ycf34 crtB, pys rbcR ndhF ndhD ndhE ndhG ndhI ndhA gltA trpB purE menA gshB prfB dgkA pabA argS nadC spoT rluD cbbK, pgk murG hisC pyrD cyanobacterial conserved hypothetical putative DNA repair protein RadA two-component response regulator fatty acid/phospholipid synthesis protein PlsX putative 1-acyl-sn-glycerol-3-phosphate acyltransfera conserved hypothetical protein Poly A polymerase family phytoene synthases conserved hypothetical protein putative Rubisco transcriptional regulator NADH dehydrogenase I chain 5 (or L) NADH dehydrogenase I chain 4 (or M) conserved hypothetical protein conserved hypothetical protein possible transcriptional regulator NADH dehydrogenase I chain 4L or K NADH dehydrogenase I chain 6 (or J) NADH dehydrogenase I chain I (or NdhI) NADH dehydrogenase I chain 1 (or H) citrate synthase Tryptophan synthase, beta chain:Pyridoxal-5'-pho Phosphoribosylaminoimidazole carboxylase Possible magnesium protoprophyrin IX methyltra two-component response regulator NifS-like aminotransferase class-V conserved hypothetical protein 1,4-dihydroxy-2-naphthoate (DHNA) octaprenyltrans putative glutathione synthetase peptide chain release factor RF-2 conserved hypothetical protein possible diacylglycerol kinase para-aminobenzoate synthase component II conserved hypothetical protein arginyl-tRNA synthetase Nicotinate-nucleotide pyrophosphorylase:Quinolinate putative thiophen / furan oxidation protein guanosine-3',5'-bis(diphosphate) 3'-diphosphatase putative pseudouridylate synthase specific to ribos conserved hypothetical protein phosphoglycerate kinase Undecaprenyl-PP-MurNAc-pentapeptide-UDPGlcNA Aminotransferases class-I Dihydroorotate dehydrogenase Table 2 ort577 ort575 ort576 ort477 ort348 ort227 ort018 ort310 ort371 ort209 ort010 ort375 ort374 ort137 ort600 ort377 ort260 ort675 ort195 ort138 ort205 ort376 ort028 ort549 ort214 ort311 ort507 ort140 ort342 ort024 ort141 ort618 ort016 ort451 ort672 ort559 ort651 Pag 2.5367 4.3721 0.9899 2.8070 3.8010 6.1653 5.7029 8.2410 3.6018 3.7407 3.8672 4.4045 5.6959 4.3567 5.3708 3.1755 9.2830 3.9285 7.1840 7.8596 6.1241 5.7757 3.5136 2.7609 7.6399 5.6999 4.8985 8.4568 1.7110 6.9624 8.7828 4.8520 3.1965 6.6821 2.5809 13.9041 3.0887 1 1 1 1 1 0 0 0 1 1 1 0 0 0 0 1 0 1 0 0 0 0 1 1 0 0 0 0 1 0 0 1 1 0 1 0 1 alr5303 alr5302 alr5300 alr5299 all3538 alr4515 alr2073 all1754 alr1396 alr3402 alr2418 all4607 all4608 alr1600 all0579 alr3561 all1717 all1318 all2473 all5037 alr0512 all4609 all2436 alr5000 alr5095 alr0520 alr3593 all1610 alr1742 alr2057 all4804 all4802 alr4798 all4316 alr3716 alr4961 all2072 glr1602 glr1601 glr1599 glr0830 gll2121 glr4178 glr4034 glr2134 gll2342 glr0710 gll2348 glr0246 glr3531 glr3015 gll2632 gll0881 glr0727 glr2919 glr0783 glr2334 gll2594 glr3530 gll1928 glr1524 gll3037 glr2580 gll1501 gll3147 glr4264 gll3278 glr3297 gsl0434 glr2933 gll3738 glr4236 gll0271 gll1463 Online Supporting Information 0201 0202 0204 0205 0208 0209 0050 0049 0048 0046 0044 1668 1669 1670 1673 1675 0659 1682 1030 1001 1684 1687 1688 1689 1691 1694 1696 1698 1704 1705 0654 1706 1707 1709 1712 1713 1716 2091 2090 2088 2087 2083 2082 2135 2136 2139 2148 2157 2169 2170 2171 2175 2177 2184 2191 2208 1386 2211 2215 2216 2219 2222 2237 2245 2247 2255 2257 2258 2259 2261 2264 2270 2271 2274 411.3 411.2 354.24 354.23 484.29 509.255 363.8 444.19 363.7 506.28 421.16 504.31 504.30 462.66 497.165 428.31 459.8 453.50 465.20 457.38 474.55 504.29 484.71 489.91 475.87 439.48 479.95 483.38 421.19 454.58 493.57 493.58 493.101 452.50 457.64 299.2 472.49 127.338 127.337 127.334 127.333 127.371 132.1045 135.2038 136.2706 136.2626 129.618 132.1049 135.1874 135.1873 129.564 129.579 125.166 136.2378 136.2871 132.1092 132.999 133.1320 131.813 128.447 135.2004 128.461 136.2535 135.1613 134.1443 129.523 129.634 136.2427 136.2526 136.2576 136.2444 132.1076 135.1745 136.2571 0227 0228 0230 0231 0235 0236 0054 0053 0052 0050 0048 1829 1830 1831 1834 1836 1841 1844 1502 1187 1847 1851 1852 1853 1855 1858 1862 1864 1871 1872 1873 1874 1875 1878 1881 1882 1885 sll1746 sll1745 sll1743 sll1742 slr0752 sll1770 sll1883 slr0553 sll1435 sll1852 sll0362 slr0293 slr0879 slr0580 sll1244 sll0202 sll1209 slr0557 sll1937 sll0157 sll1005 sll0171 slr1720 sll1443 slr0527 sll0378 sll0290 slr1652 sll0170 slr1559 sll1737 sll1767 slr0585 sll0657 slr1844 sll1520 sll1172 tlr0298 tlr0297 tlr0295 tlr0294 tlr0658 tlr2242 tll1911 tlr0898 tlr2394 tlr0267 tll2103 tll1603 tlr1678 tlr2020 tlr2413 tlr0535 tll0857 tlr1503 tlr0192 tlr2433 tlr0809 tll0745 tll1144 tll0768 tlr1732 tlr0144 tlr0213 tll2039 tlr1733 tll0590 tlr1073 tlr1279 tlr0712 tll0971 tll0184 tll1899 tll1227 121.5561 121.5560 121.5558 121.5557 121.6150 121.5999 122.7762 116.2769 122.6707 101.113 120.4504 118.3757 118.3755 103.292 103.305 121.6104 112.1647 121.5381 110.1285 122.6983 118.3754 114.2154 115.2408 104.403 119.4300 101.169 108.973 118.3658 112.1833 109.1223 120.4830 120.4831 114.2209 122.7016 121.5477 118.3637 121.6077 2340 2341 2343 2344 2348 2349 2353 2354 2356 2358 2360 2374 2375 2376 2378 2382 2389 2396 2401 2402 2418 2425 2426 2428 2431 2475 2495 2497 2508 2509 2510 2511 2513 2516 2522 2523 2526 232.1507 232.1506 232.1504 232.1503 232.2417 232.2284 232.2033 232.4174 232.2836 232.2327 232.4120 232.1698 232.1697 232.3083 232.4219 232.2439 232.4204 232.1906 232.4292 232.1257 232.1842 232.1696 232.4140 232.1228 232.1322 232.1848 232.2076 232.3091 232.4186 232.2021 232.1045 232.1043 232.1039 232.5124 232.2490 232.1198 232.2032 241.1484 241.1483 188.861 188.860 243.1513 318.2921 225.1272 357.4421 317.2913 362.6188 131.388 314.2812 358.4606 233.1366 330.3277 349.3904 361.5402 339.3526 361.5768 328.3193 354.4155 254.1664 353.4089 88.174 362.6090 361.5933 362.6491 342.3653 362.6498 336.3438 361.5451 361.5452 141.464 360.5013 314.2814 295.2380 362.6189 rpl12 rpl10 rpl11 nusG eno argJ coaE gatB alaS gcvP gcsH rpl9 gidA valS gcvT aspS pyrG cobA ppk dnaK2 aroE ycf60 rps6 argG mraY uvrA recN thrC 50S ribosomal protein L7/L12 50S ribosomal protein L10 50S ribosomal protein L11 transcription antitermination protein, NusG Enolase possible kinase glutamate N-acetyltransferase / ornithine N-acetyltran putative dephospho-CoA kinase glutamyl-tRNA (Gln) amidotransferase subunit B Nucleoside diphosphate kinase Alanyl-tRNA synthetase Glycine cleavage system P-protein putative Glycine cleavage H-protein conserved hypothetical protein 50S ribosomal protein L9 glucose inhibited division protein A Putative DNA ligase valyl-tRNA synthetase Ferric uptake regulator family conserved hypothetical protein MazG family protein putative Glycine cleavage T-protein (aminomethyl tra Aspartyl-tRNA synthetase:GAD domain:tRNA syn Glutamine amidotransferase class-I:CTP synthase possible ATPase putative uroporphyrin-III c-methyltransferase putative polyphosphate kinase conserved hypothetical protein Molecular chaperone DnaK2, heat shock protein h Shikimate / quinate 5-dehydrogenase conserved hypothetical protein 30S ribosomal protein S6 Argininosuccinate synthase Putative phospho-N-acetylmuramoyl-pentapeptide-tra excinuclease ABC subunit A putative DNA repair protein threonine synthase Figure legends Page 19 Fig. 6. Representative backbone tree topologies and other hypotheses used in the ShimodairaHasegawa test. In order to determine which of the above topologies best represent the evolutionary history of the conserved protein families, we performed a Shimodaira-Hasegawa (SH) test (74) in which the fitness of a particular topology to the 682 sequence alignments was analyzed. To aid the comparison, we also generated topologies that display alternative placement of SCO and TEL with minor or no changes in topologies of the conserved clades (T6-T10), as well as topologies that break the conserved clades at random (T11-T15). Note that only 2~3% topological divergence is displayed. Species abbreviations as in Table 1. Fig. 7. Compatibility of tree topologies to the orthologous datasets. The SH tests show the number of datasets accepting (black bars) or rejecting (grey bars) each of the 15 topologies at the 5% significance level. The SH tests indicated that almost all (97.5% to 99.6%) of the datasets support topologies T1-T5 at the 95% confidence level (P=0.95), whereas much fewer datasets support topologies T6-T11, and T11-T15 were strongly rejected by most of the datasets. While this analysis corroborated the main stream as discerned through genomic approaches, it was inadequate in choosing a unique, preferable topology to other similar ones. This result suggests that artificial paralogs or genes potentially obtained by LGT may thwart the establishment of organismal relationships based on plural consensus of individual gene phylogenies. Online Supporting Information Figure 6 Page 20 SSU rRNA Concatenated proteins 16S_NJ/ML/MP; NJ_γ NJD; NJJ; NJK;QP ANA AVA NPU TER CWA SYN PMM PMS PMT SYW SCO TEL GVI T1 ANA AVA NPU TER CWA SYN TEL PMM PMS PMT SYW SCO GVI T2 Consensus and supertree ProtML; ML_consensus; ML_supertree ANA AVA NPU TER CWA SYN PMM PMS PMT SYW SCO TEL GVI T3 NJ_consensus NJ_supertree ANA AVA NPU TER CWA SYN TEL SCO SYW PMT PMS PMM GVI T4 ANA AVA NPU TER CWA SYN TEL SYW PMT PMS PMM SCO GVI T5 Other hypotheses SYW PMT PMS PMM ANA AVA NPU SYN CWA SCO TER TEL GVI T6 ANA AVA NPU TER CWA SYN SCO TEL PMM PMS PMT SYW GVI T7 SYW PMT PMM PMS SCO TEL SYN NPU AVA ANA CWA TER GVI T11 ANA AVA NPU TER CWA SYN SCO SYW PMT PMS PMM TEL GVI T8 SYN NPU ANA AVA TER CWA SCO TEL PMT PMM PMS SYW GVI T12 ANA AVA NPU TER CWA SYN TEL SCO PMM PMS PMT SYW GVI T9 ANA AVA NPU TEL CWA SYN SYW PMM PMT SCO PMS TER GVI T13 SYW PMS PMT PMM SCO SYN CWA ANA AVA NPU TEL TER GVI T10 ANA AVA NPU CWA TER PMT PMS PMM SYW SCO TEL SYN GVI T14 CWA SCO SYN PMT PMS SYW ANA AVA NPU TER TEL PMM GVI T15 Shi & Falkowski, SI Fig. 6 Online Supporting Information Figure 7 Page 21 700 Number of orthologs 650 600 550 500 150 100 50 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 Tree topologies Shi & Falkowski, SI Fig. 7 Online Supporting Information