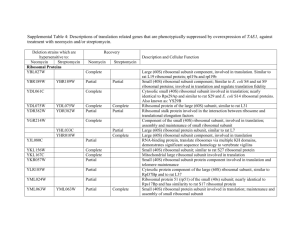
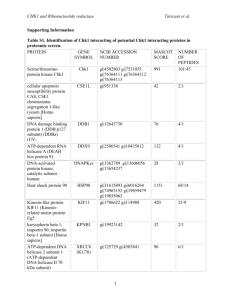
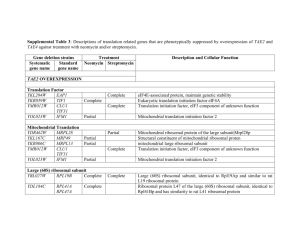
Table 5s: Genes down-regulated by both ethanol and n
advertisement

Table 5s: Genes down-regulated by both ethanol and n-butanol. Systematic name YJR047C YKR061W YLL034C YHR209W YIL117C YOR340C YHL047C YMR194W YJR070C YAL059W YJL109C YDL201W YPL198W YHR030C YHR128W YIL066C YPL012W YLR196W YOR009W YLR022C YMR238W YDR101C YKR099W YLR223C YKR091W YOR080W YPL226W YJL033W YOL041C YGR189C YNL132W Description translation initiation factor eIF-5A, anaerobically expressed form mannosyltransferase (putative)|type 2 membrane protein Putative ATPase of the AAA family, required for export of pre-ribosomal large subunits from the nucleus; distributed between the nucleolus, nucleoplasm, and nuclear periphery depending on growth conditions Putative S-adenosylmethionine-dependent methyltransferase of the seven beta-strand family Pheromone-regulated protein, predicted to have 1 transmembrane segment; induced during cell integrity signalling DNA dependent RNA polymerase I subunit A43 triacetylfusarinine C transporter ribosomal protein L36A (L39) (YL39) Protein that binds to the C-terminal domain of Hyp2p (eIF5A); has four to five HEAT-like repeats Protein of unknown function, localized in the nucleoplasm and the nucleolus, genetically interacts with MTR2 in 60S ribosomal protein subunit export U3 snoRNP protein Subunit of a tRNA methyltransferase complex composed of Trm8p and Trm82p that catalyzes 7-methylguanosine modification of tRNA ribosomal protein L7B (L6B) (rp11) (YL8) Suppressor of lyt2 UPRTase ribonucleotide reductase, large (R1) subunit Required for normal pre-rRNA Processing. Member of a group of seven genes whose expression is repressed during growth on glucose before and during the diauxic shift. Protein with periodic trytophan residues that resembles members of betatransducin superfamily because of presence of WD-40 repeats cell wall mannoprotein Protein required for cell viability Mannosidase, essential glycosylphosphatidylinositol (GPI)-anchored membrane protein required for cell wall biogenesis, involved in filamentous growth, homologous to Dcw1p --transcription factor Essential protein with a highly acidic N-terminal domain; IFH1 exhibits genetic interactions with FHL1, overexpression interferes with silencing at telomeres and HM loci; potential Cdc28p substrate Cytoplasmic protein that, when overexpressed, suppresses the lethality of a rad53 null mutation; potential Cdc28p substrate Protein of unknown function, involved in invasive and pseudohyphal growth This gene encodes a protein with an Q/N-rich amino terminal domain that acts as a prion, termed [NU]+. RNA helicase (putative) Nucleolar protein, required for pre-25S rRNA processing; contains an RNA recognition motif (RRM) and has similarity to Nop13p, Nsr1p, and putative orthologs in Drosophila and S. pombe cell wall protein Killer toxin REsistant Stndard name ANB1 KTR2 ethanol fold change 9.943508 6.931792 butanol fold change 49.91542 3.938259 RIX7 6.929144 29.15893 6.35241 4.25416 PRM5 RPA43 ARN2 RPL36A 6.336404 6.048 5.33824 5.31805 16.17019 11.90551 5.955199 9.783639 LIA1 5.270697 11.49286 ECM1 UTP10 5.205433 5.107464 6.110613 14.92343 TRM8 RPL7B SLT2 FUR1 RNR3 5.097241 5.041165 4.991965 4.990953 4.771885 19.86828 13.29336 4.821625 5.685295 12.73316 RRP12 4.702785 11.77186 PWP1 TIR4 SDO1 4.692346 4.669574 4.525581 14.89848 3.548258 4.914141 DFG5 ARX1 BAS1 4.446409 4.417974 4.391437 4.73233 16.38624 5.012216 IFH1 4.287206 3.748858 SRL3 DIA2 4.229859 4.204004 6.05013 7.57732 NEW1 HCA4 4.177521 4.141871 11.42627 8.037202 NOP12 CRH1 KRE33 4.077821 4.061059 4.033654 9.980952 8.127182 11.90071 YHR209W YLR175W YJR094W-A YBL004W YLR009W YLR432W YLR449W YHL040C YLR222C YKL172W YOR104W YGL078C YNL110C YKL161C YKR060W YFL034C-A YLR367W YGR081C YLL045C YDR184C YBR247C YLR060W YBR143C YAL025C YNR054C YLR355C YMR309C YHR089C YIL133C YMR049C YHR142W YPL082C YPL051W YKR026C YDR190C YPR010C YMR128W YKL009W YML022W YLR406C YGR245C YJL010C YKL082C YKR024C YPR112C YDL166C major low affinity 55 kDa centromere/microtubule binding protein ribosomal protein L43B U3 snoRNP protein part of a pre-60S complex IMP dehydrogenase homolog peptidyl-prolyl cis-trans isomerase (PPIase) Transporter, member of the ARN family of transporters that specifically recognize siderophore-iron chelates; responsible for uptake of iron bound to ferrirubin, ferrirhodin, and related siderophores U3 snoRNP protein nucleolar protein [PSI+] induction ATP dependent RNA helicase|dead/deah box protein CA3 ribosome biogenesis Mpk1-like protein kinase; associates with Rlm1p Possible U3 snoRNP protein involved in maturation of pre-18S rRNA, based on computational analysis of large-scale protein-protein interaction data ribosomal protein L22B (L1c) (rp4) (YL31) ribosomal protein S22B (S24B) (rp50) (YS22) Protein of unknown function; deletion mutant has synthetic fitness defect with an sgs1 deletion mutant ribosomal protein L8B (L4B) (rp6) (YL5) Nuclear protein, possibly involved in regulation of cation stress responses and/or in the establishment of bipolar budding pattern 57 kDa protein with an apparent MW of 70 kDa by SDS-PAGE (putative) phenylalanine-tRNA ligase subunit eRF1 (eukaryotic Release Factor 1) homolog nuclear protein (putative) Protein required for cell viability acetohydroxyacid reductoisomerase translation initiation factor eIF3 subunit small nucleolar RNP protein ribosomal protein L16A (L21A) (rp22) (YL15) Protein required for maturation of the 25S and 5.8S ribosomal RNAs; homologous to mammalian Bop1 The seventh gene identified that is involved in chitin synthesis; involved in Chs3p export from the ER helicase (putative) Similar to ADP-ribosylation factor. Part of the carboxypeptidase Y pathway. eIF2B 34 kDa alpha subunit RUVB-like protein, TIP49a Homologue RNA polymerase I subunit U3 snoRNP protein Protein involved in mRNA turnover and ribosome assembly, localizes to the nucleolus adenine phosphoribosyltransferase ribosomal protein L31B (L34B) (YL28) Severe Depolymerization of Actin Protein required for cell viability Required for normal pre-rRNA Processing RNA helicase (putative) Essential conserved protein that associates with 35S precursor rRNA and is required for its initial processing at the A(0)-A(2) cleavage sites, shows partial nucleolar localization, contains five consensus RNA-binding domains Essential nuclear protein, involved in the oxidative stress response CBF5 RPL43B UTP20 RLP24 IMD3 FPR4 4.010898 3.963027 3.961827 3.961351 3.912019 3.815077 15.85436 5.418309 8.144909 13.00972 4.119831 12.62768 ARN1 UTP13 EBP2 PIN2 DBP3 NOP15 YKL161C 3.788784 3.784244 3.781773 3.755538 3.755048 3.751731 3.735409 8.488774 18.41796 5.89372 4.179876 8.488889 8.250732 5.079365 UTP30 RPL22B RPS22B 3.72327 3.717791 3.681528 16.44444 4.263852 11.25726 SLX9 RPL8B 3.679775 3.673766 5.261044 12.54299 ATC1 ENP1 FRS1 SUP45 MAK16 ESF2 ILV5 NIP1 GAR1 RPL16A 3.660759 3.635336 3.621895 3.619587 3.597917 3.593828 3.589816 3.51511 3.5 3.499824 4.32287 7.296454 8.816184 5.54954 6.853175 11.09138 9.124553 7.198312 9.029353 7.735518 ERB1 3.477925 14.25384 CHS7 MOT1 ARL3 GCN3 RVB1 RPA135 ECM16 3.466276 3.440488 3.436317 3.396083 3.395376 3.392757 3.392543 5.316342 3.745293 4.947955 7.101251 8.001932 8.787796 7.044086 MRT4 APT1 RPL31B SDA1 YJL010C RRP14 DBP7 3.386161 3.378409 3.368905 3.358696 3.311419 3.311255 3.288486 7.901042 6.061162 7.425894 5.421053 8.394737 10.33041 7.993711 MRD1 FAP7 3.280593 3.277108 13.20398 3.439928 YPL217C YKL143W YER145C YLR130C YDR441C YLR061W YDL075W YPR163C YLR106C YFR001W YMR319C YML055W YLR265C YER045C YMR229C YLR401C YOR216C YDR496C YPL093W YJR002W YHR148W YPR143W YLL011W YNR053C YKL006W YGR151C GTP-binding protein required for processing of 35S pre-rRNA at sites A0, Protein required for viability at low temperature iron permease low affinity zinc transport protein Apparent pseudogene, not transcribed or translated under normal conditions; encodes a protein with similarity to adenine phosphoribosyltransferase, but artificially expressed protein exhibits no enzymatic activity ribosomal protein L22A (L1c) (rp4) (YL31) ribosomal protein L31A (L34A) (YL28) /// ribosomal protein L31B (L34B) (YL28) translation initiation factor eIF-4B midasin Nuclear protein involved in asymmetric localization of ASH1 mRNA; binds double-stranded RNA in vitro --signal peptidase complex subunit|similar to mammalian protein SPC25 Mating-type regulated component of NHEJ Basic leucine zipper (bZIP) transcription factor of the ATF/CREB family, may regulate transcription of genes involved in utilization of non-optimal carbon sources U3 snoRNP protein dihydrouridine synthase 3 Novel matrix protein that is involved in the structural organization of the cisGolgi. Relieves uso1-1 transport defect; golgin-160 related protein. member of the PUF protein family homologs identified in human and Trypanosoma brucei|nucleolar G-protein (putative) U3 snoRNP protein U3 snoRNP protein Protein required for cell viability U3 snoRNP protein part of a pre-60S complex ribosomal protein L14A Gtp-binding protein of the ras superfamily involved in bud site selection BMS1 LTV1 FTR1 ZRT2 3.262921 3.256437 3.240099 3.234745 5.808 8.448898 3.62963 5.513498 APT2 RPL22A 3.221938 3.216722 3.863857 43.12709 RPL31A TIF3 MDN1 3.211286 3.208214 3.193151 11.04741 10.43603 3.569678 LOC1 FET4 SPC2 NEJ1 3.189213 3.153161 3.151712 3.149938 7.875644 14.83039 9.126496 5.502165 ACA1 RRP5 DUS3 3.133333 3.129923 3.107492 4.049352 8.898327 5.088 RUD3 3.07277 6.14554 PUF6 3.059524 9.388128 NOG1 MPP10 IMP3 RRP15 SOF1 NOG2 RPL14A RSR1 3.059322 3.057872 3.053435 3.052469 3.044232 3.029573 3.011876 3.002744 7.18408 3.49174 6.060606 4.654118 8.552477 8.684226 4.862958 4.908527