Supplementary Methods and Procedures.
advertisement

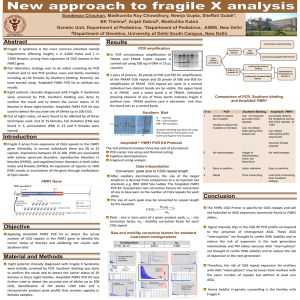
SUPPLEMENTARY MATERIALS AND METHODS Molecular Measures Blood was collected in EDTA vacutainers, and genomic DNA (gDNA) isolated from 5 ml of peripheral blood leukocytes using Gentra Puregene Blood Kit (Qiagen, Germantown, MD, USA). The gDNA concentration was measured using the Nanodrop Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Southern blot and PCR analysis were conducted as previously described.1,2 CGG size was calculated using FluorChem 8900 software (Alpha Innotech, San Leandro, CA, USA). To determine the presence and locations of the AGG interruptions within each FMR1 allele, PCR was performed using 100 ng of gDNA, and a combination of a two-tier PCR approach using a CGG linker primer as previously described1 and the Human FMR1 PCR kit (Asuragen Inc., Austin, TX, USA), based on manufacturer recommendations and as previously described. 3,4 The latter was performed in 15 µl final reaction volume. DNA was amplified using the ABI GeneAmp 9700 thermal cycler (Applied Biosystems, Carlsbad, CA, USA) under the following conditions: 95°C for 5 minutes; followed by 10 cycles of 97°C for 35 seconds, 62°C for 35 seconds 68°C for 4 minutes; 20 cycles of 97°C for 35 seconds, 62°C for 35 seconds 68°C for 4 minutes and an additional 20 seconds per cycle; 72°C extension for 10 minutes and finally incubation at 4°C. Details are as previously described.1,3 Electrophoresis Two microliters of PCR product were denatured with 2 µl of ROX 1000 size marker (Asuragen Inc.) in Hi-Di Formamide (Applied Biosystems) to a total volume of 15 µl for 2 minutes at 95°C, followed by 3 minutes at 4°C. The singlestranded FAM-6 labeled PCR product was detected by capillary electrophoresis using the ABI 3100 Genetic Analyzer (Applied Biosystems). Fragments were separated through a 36-cm capillary array using POP4 polymer (Applied Biosystems) following manufacturer’s recommendations with the following changes to default run parameters: injection for 20 seconds at 1,250 volts, and a run time of 50 minutes. Determination of AGG Interruptions Genescan files were analyzed using Peakscanner Software (v1.0) (Applied Biosystems). The full fragment band(s) amplified from the C and F primers were first identified, the size of which was determined by comparing to ROX1000 size standard (Asuragen Inc.) and using an in-house DNA ladder comprising PCR alleles previously sequenced (20, 29, 30, 31, 55, 75, 88, and 108 CGG repeats in length). The remaining peaks (here referred to as CGG peaks) mark the presence of CGG repeats in the amplicon, each being the result of the CGG linker primer annealing to a CGG5 region and elongating towards the F primer. In females, the CGG peak can occur from amplification of either of the two X chromosomes. The first CGG peak detected by electropherogram corresponds to the fifth CGG unit from the F primer, the second peak detected corresponds to the sixth CGG unit, and so on. The presence of a CGG peak that is twice the height (intensity) of a neighboring peak is the result of amplification on the FMR1 allele on both chromosomes. The presence of one CGG peak and one AGG interruption at the same location (female) will result in a peak with the intensity expected for one allele (half the height of the previously described peak). Likewise, the absence of a CGG repeat and the presence of either one (male) or two (female) AGG interruptions results in a decrease in CGG peak intensity to baseline levels. The presence of CGG peaks and their corresponding height thus allows the entire locus to be characterized so that the location of CGG and AGG segments are precisely determined. Using this assay, the initial scanned fluorescence signal corresponds to the 5’ end of the amplicon (details are as previously described).3,4 The total CGG length is characterized as the number of CGG repeats, plus AGG interruptions if present; and corresponds to the CGG length reported out by PCR analysis. The pure CGG stretch is characterized as the longest segment of repeating CGG units within the total CGG length that does not contain an interrupting AGG. In premutation alleles, the longest pure CGG stretch is characteristically on the 3’ end of the microsatellite locus, with AGG interruptions typically occurring within the first 9 to 32 trinucleotide units. 5 Eci I Digestion The presence of AGG interruptions was confirmed in a subset of samples by digestion with the EciI restriction enzyme (New England Biolabs Inc., Ipswich, MA, USA). The recognition site for EciI cuts 11 nucleotides after the adenine of the AGG interruption (5’-GGCGGA (N)11 -3’).6 For this assay, gene-specific PCR was performed using the Human FMR1 PCR kit without the addition of the CGG linker primer, and using the following PCR thermal cycler conditions (15 µl total volume): 98°C for 5 minutes followed by 25 cycles of 97°C for 35 seconds, 62°C for 35 seconds, 72° for 4 minutes; a final extension was performed at 72°C for 10 minutes, followed by short-term storage at 4°C. Following PCR amplification, 10 µl of PCR product was digested with 0.6 ul EciI (New England Biolabs) in 20 µl total reaction volume for 4 hours at 37°C, followed by heat inactivation of the enzyme for 20 minutes at 67°C. Both digested and undigested samples were run on an agarose gel using 1X Bionic buffer (Sigma Aldrich, St. Louis, MO, USA) for improved resolution.2 Fragments were separated for 15 minutes at 80 volts followed by 70 minutes at 150 volts. The gel was stained using Sybr Gold (Invitrogen, Carlsbad, CA, USA) and visualized with the FluorChem 8900 (Alpha Innotech). Size ranges were determined and compared to the results from capillary electrophoresis separation for data consistency (Supplementary Figure 1). Haplotype Markers Four markers flanking the FMR1 microsatellite were analyzed in premutation carrier mothers and offspring, as necessary and with sample availability: DXS548, FRAXAC1, ATL1 (rs4949), and IVS10 (rs25714). The DXS548 and FRAXAC1 loci were amplified as described7,8 with minor modifications. Briefly, PCR was performed using 100 ng of gDNA, Amplitaq Gold polymerase with Buffer II (Applied Biosystems), 2.5mM MgCl, 200 M of each dNTP (Roche, Indianapolis, IN, USA), 0.2 M of each primer (DXS548: (forward) 5’ 6-FAM-GAA TAG TCT CTG GGG TGG ATC TC 3’; (reverse) 5’ AGA GCT TCA CTA TGC AAT GGA ATC 3’) and (FRAXAC1: (forward) 5’ 6- FAM-GAT CTA ATC AAC ATC TAT AGA CTT TAT T 3’; (reverse) 5’ GAT GAG AGT CAC TTG AAG CTG G 3’). Thermal cycling conditions for DXS548 and FRAXAC1 were as follows: 95C for 7.5 minutes, 10 cycles of 98C for 30 seconds, 60C for 1.5 minutes, 72C for 1.5 minutes; 22 cycles of 98C for 30 seconds, 55C for 1.5 minutes, 72C for 1.5 minutes; and a final extension for 10 minutes at 72C. SNPs ATL1 and IVS10 were amplified using Taqman Universal Master Mix according to manufacturer’s recommendations. FRAXAC1 and DXS548 PCR products were separated using capillary electrophoresis on an ABI 3100 Genetic Analyzer (Applied Biosystems) using standard run parameters and POP7 polymerase with LIZ 500 size standard (Applied Biosystems). Raw data was analyzed using Peakscanner (Applied Biosystems). ATL1 and IVS10+14 were analyzed using the ABI 7900HT with SDS software (Applied Biosystems). Statistical Methods The association of SNPs rs4949 and rs25714 with maternal total CGG repeat number or pure CGG stretch was analyzed using ANOVA models, using one observation from each mother. The association of each SNP with number of maternal AGG interruptions was analyzed using Fisher's exact test. All statistical analyses were conducted using R, version 2.13.0.9 The association between haplotype and interspersion pattern was analyzed using chi-square tests, with p-values calculated via Monte Carlo simulation. _ENREF_10SUPPLEMENTARY REFERENCES 1. Tassone F, Pan R, Amiri K, Taylor AK, Hagerman PJ. A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J Mol Diagn. Jan 2008;10(1):43-49. 2. Filipovic-Sadic S, Sah S, Chen L, et al. A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clin Chem. Mar 2010;56(3):399-408. 3. Chen L, Hadd A, Sah S, et al. An information-rich CGG repeat primed PCR that detects the full range of fragile X expanded alleles and minimizes the need for southern blot analysis. J Mol Diagn. Sep 2010;12(5):589-600. 4. Yrigollen CM, Tassone F, Durbin-Johnson B, Tassone F. The role of AGG interruptions in the transcription of FMR1 premutation alleles. PLoS One. 2011;6(7):e21728. 5. Eichler EE, Hammond HA, Macpherson JN, Ward PA, Nelson DL. Population survey of the human FMR1 CGG repeat substructure suggests biased polarity for the loss of AGG interruptions. Hum Mol Genet. Dec 1995;4(12):2199-2208. 6. Tassone F, Beilina A, Carosi C, et al. Elevated FMR1 mRNA in premutation carriers is due to increased transcription. RNA. Apr 2007;13(4):555-562. 7. Riggins GJ, Sherman SL, Oostra BA, et al. Characterization of a highly polymorphic dinucleotide repeat 150 KB proximal to the fragile X site. Am J Med Genet. Apr 15-May 1 1992;43(1-2):237-243. 8. Zhong N, Dobkin C, Brown WT. A complex mutable polymorphism located within the fragile X gene. Nat Genet. Nov 1993;5(3):248-253. 9. R: A language and environment for statistical computing. [computer program]. Version 2.13.0. Vienna, Austria: R foundation for Statistical Computing; 2011.