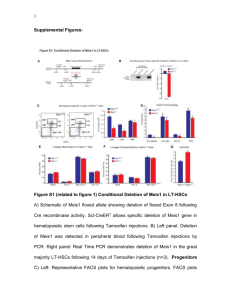
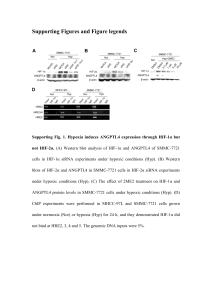
Jeong et al. Supplemental Data HIF
advertisement

Jeong et al. Supplemental Data HIF-1α immunoassay Methods for specimen collection and processing to preserve HIF-1α in 18-gauge tumor needle biopsies have been validated and a modified version of the DUOSet IC ELISA Kit from R&D Systems was validated to quantitatively measure HIF-1α levels in human tissue biopsy [1]. Core needle biopsies (18-gauge) were immediately dry flash frozen and stored at -80°C. At the time of the assay, samples were thawed in 250 µL nitrogen-purged extraction buffer containing protease inhibitors (Roche Applied Science), phenylmethylsulfonyl fluoride (Sigma-Aldrich), and the prolyl hydroxylase inhibitor 2-hydroxyglutarate (3B Scientific Corporation), which stabilizes HIF-1 protein. Cells were lysed using Precellys 2.8 mm ceramic bead tubes and the Precellys24 tissue homogenizer; protein content was determined using the BCA protein assay kit (Thermo Scientific). A minimum lysate concentration of 0.5µg/µL was needed to proceed with the assay. For the immunoassay, 100 µL of DUOSet capture antibody at a concentration of 4 µg/mL (Lot# KQE0411012) in 1x PBS was added to each well of a 96-well white microtiter plate and incubated at 2°C-8°C for 16±1 h. Wells were blocked with 300 µL Reagent Diluent (1x PBS, 0.05% Tween 20, 5% bovine serum albumin) at 24°C for 1-2 h. DUOSet HIF-1α standard was serially diluted in Reagent Diluent to a range of 7.8 to 1000pg HIF-1α/mL and served as standard controls. HIF-1α standards or controls were loaded in 100 µL total volume with Reagent Diluent; clinical samples were loaded in triplicate at 5, 7.5, and 10 µL cell extract diluted in Reagent Diluent per well onto each plate and incubated at 2°C-8°C for 16±1 h. Next, 100 µL/well of DUOSet HIF-1α detection antibody (100 ng/mL, Lot# KKR0311012) in Reagent Diluent was added and incubated at 24°C for 2 h. Then 100 µL/well DUOSet streptavidinhorseradish peroxidase conjugate (KPL, Gaithersburg, MD) at a final concentration of 5 µg/mL Jeong et al. (1:200, Lot# 1260918) in Reagent Diluent was added and incubated at 24°C for 30 min. Finally, 100 µL/well of room temperature LumiGLO Chemiluminescent Substrate (KPL) was added and the plate immediately read on a Tecan Infinite M200 plate reader (Tecan Systems, San Jose, CA). Relative light unit values were plotted using a HIF-1α analysis template to generate standard curves. Average HIF-1α level, standard deviation, and CV for each tumor extract were determined from the HIF-1α standard curve. Final HIF-1α readout for each sample was reported as pg HIF-1α per 1 μg total protein using the HIF-1α standard curve. RNA extraction and real-time PCR analysis After collection, tumor biopsy samples were submerged in RNAlater RNA Stabilization Reagent (Qiagen), kept at 4°C for 24 hours, and then stored at -20°C. Samples were subsequently disrupted and homogenized in RNA Lysis Buffer (RNeasy Mini Kit, Qiagen) using FastPrep (Lysing Matrix D columns, MP Biomedicals). Total RNA was extracted according to the manufacturer's procedure. Genomic DNA was digested using RNase-Free DNAse Set (Qiagen) during RNA extraction. RNA integrity was evaluated using RNA 6000 Nano Assay in an Agilent 2100 Bioanalyzer (Agilent Technologies). RT-PCR was carried out using an RT-PCR kit (PE Biosystems) according to the manufacturer's procedure. Expression of VEGF, GLUT-1, HK2, PDK1 and CA9 were assessed by real-time PCR using a 7500 Real-Time PCR System (Applied Biosystems). Typically, 5 ng of reverse-transcribed cDNA per sample was used to conduct real-time PCR in triplicate samples. Primers and probes used are listed below in Supplementary Table S1. Detection of 18S rRNA, used as internal control, was carried out using premixed reagents from Applied Biosystems. Detection of VEGF and 18S rRNA was carried out using TaqMan Universal PCR Master Mix (Applied Biosystems), whereas GLUT-1, HK2, Jeong et al. PDK1 and CA9 detection was carried out using Sybr Green PCR Master Mix (Applied Biosystems). Values are expressed as percent change relative to pretreatment samples for each patient. HIF-1α and B2M mRNA copy numbers in tumor tissues were determined in comparison with standard curves constructed using 30, 3x102, 3x103, 3x104 and 3x105 copies of expression plasmids molecules. HIF-1α mRNA copy numbers were normalized to mRNA copy numbers of the house keeping gene B2M and expressed as HIF-1α/B2M copy number. A pShuttle-HIF-1α expression vector was generated by inserting human full-length wild type HIF-1α cDNA into pShuttle (Clontech) while the B2M expression plasmid (pBJ1-hB2M) was obtain by Addgene (plasmid 12099) [2]. DCE-MRI Technique Imaging was performed using a 1.5-T MR system (Philips Achieva, The Best, The Netherlands) with a dedicated receive-only 6-channel phased array coil. First, the diagnostic T2-weighted images, that were used to locate the target tumor, were obtained using (TR/TE) 4600/100 msec, a section thickness of 6 mm, 400 mm field of view, and a matrix of 320 x 320. Next, the unenhanced T1-weighted images, used to determine the tissue T1 map, were acquired with a three-dimensional spoiled gradient- echo sequence by using (TR/TE) 9/3.6 msec, a 5° flip angle, 5 mm-thick sections, 400 mm field of view, and a matrix of 256x256. Finally, DCE-MR images were obtained with a three-dimensional spoiled gradient- echo sequence using (TR/TE) 9/3.6msec, a 15° flip angle, 5-mm-thick sections through the entire target lesion, 400 mm field of view, an acquisition time of 30 seconds per data set, and a matrix of 256 x 256. After three baseline unenhanced image acquisitions, an automatic injector (Medrad Spectris, Indianola, PA) Jeong et al. was used to intravenously infuse gadopentetate dimeglumine (Magnevist; Bayer Healthcare Pharmaceuticals, Wayne, NJ) at 0.3 mL/sec, for a total of 0.1 mmol per kilogram of body weight (typically 15–20 mL), followed by a 50-mL normal saline flush. Continuous 30-second imaging data sets were obtained before, during, and after administration of the contrast medium for a total of 8 minutes to result in 23 repeated data sets. Patients were asked to hold their breath during MR image acquisitions. DCE-MRI Analysis Data from DCE-MRI were analyzed using a computer-based 2-compartment model based on the Kety model, which has also been termed the general kinetic model (GKM) [3]. GKM incorporates an arterial input function derived from aortic measurements and a T1 map for converting signal intensity to gadolinium concentration. It is based on a 2-compartment model that assumes that the vascular space is in rapid equilibrium with the extravascular, extracellular space and further assumes a rapid water exchange between intra- and extracellular water. GKM analysis was performed in an IDL-based (Interactive Data Language; Research Systems Inc., Boulder, CO) research tool (Cine Tool, GE Healthcare). Region-of-interest measurements were obtained from one selected target lesion in each patient. Two parameters derived from the curve fitting GKM algorithm were used to perform the quantitative analysis: Ktrans, the forward contrast transfer rate, kep, and the reverse contrast transfer rate. References 1. Park SR, Kinders RJ, Khin S, Hollingshead M, Parchment RE, Tomaszewski JE, Doroshow JH (2012)Validation and fitness testing of a quantitative immunoassay for HIF-1 alpha in biopsy specimens. Cancer Res 72: 3616 Jeong et al. 2. Olson R, Huey-Tubman KE, Dulac C, Bjorkman PJ (2005) Structure of a pheromone receptor-associated MHC molecule with an open and empty groove. PLoS Biol 3: e257 3. Kety SS (1951) The theory and applications of the exchange of inert gas at the lungs and tissues. Pharmacol Rev 3: 1-41 Jeong et al. Supplemental Table S1. RT-PCR primers. Primer name Sequence HIF-1a forward CCAGTTACGTTCCTTCGATCAGT HIF-1a reverse TTTGAGGACTTGCGCTTTCA B2M forward GACTTGTCTTTCAGCAAGGA B2M reverse ACAAAGTCACATGGTTCACA VEGF forward TACCTCCACCATGCCAAGTG VEGF reverse ATGATTCTGCCCTCCTCCTTC GLUT-1 forward GATCCTGGGCCGCTTCAT GLUT-1 reverse ACATGGGCACGAAGCCTG PDK1 forward CAAGAATGCAATGAGAGCCACTA PDK1 reverse CCAGCGTGACATGAACTTGAA CAIX forward ACATGGGCACGAAGCCTG CAIX reverse AACTGCTCATAGGCACTGTTTTCTT Jeong et al. Supplemental Table S2. HIF target genes Gene symbol Gene name Function VEGF vascular endothelial growth factor A Growth factor active in angiogenesis, vasculogenesis and endothelial cell growth PDK1 pyruvate dehydrogenase kinase, isozyme 1 Inhibits the mitochondrial pyruvate dehydrogenase complex, thus contributing to the regulation of glucose metabolism CA9 (CAIX) carbonic anhydrase IX Catalyzes the reversible hydration of carbon dioxide GLUT1 solute carrier family 2 (facilitated glucose transporter), member 1 Facilitative glucose transporter