Biology 433
advertisement
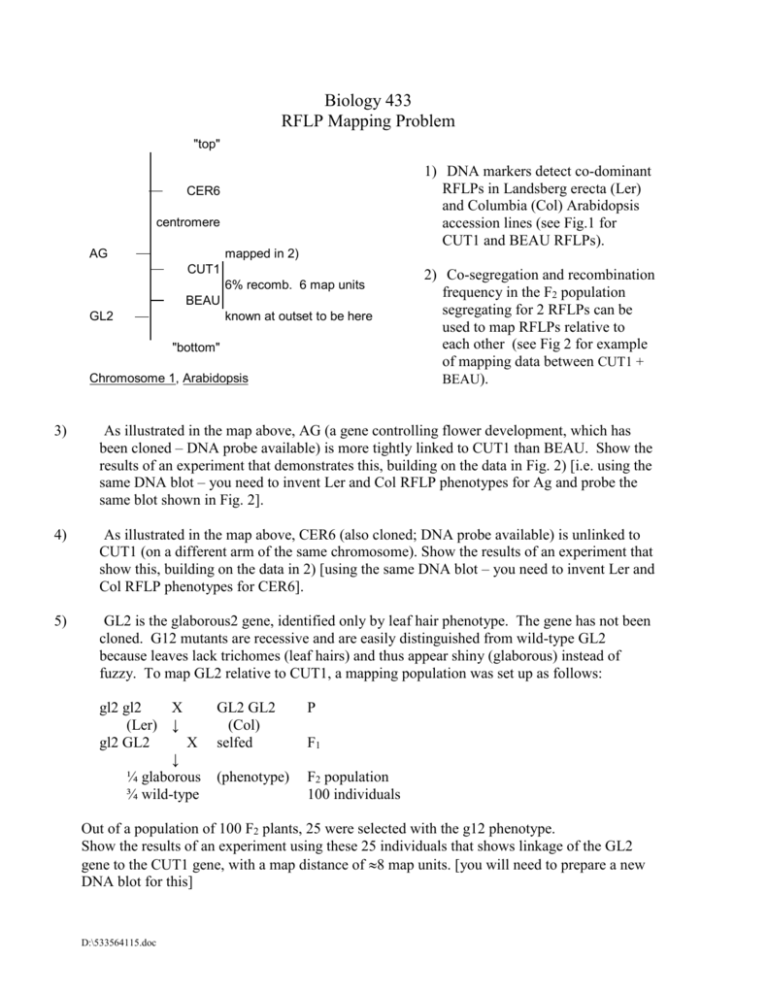
Biology 433 RFLP Mapping Problem "top" 1) DNA markers detect co-dominant RFLPs in Landsberg erecta (Ler) and Columbia (Col) Arabidopsis accession lines (see Fig.1 for CUT1 and BEAU RFLPs). CER6 centromere AG mapped in 2) CUT1 6% recomb. 6 map units BEAU GL2 known at outset to be here "bottom" Chromosome 1, Arabidopsis 2) Co-segregation and recombination frequency in the F2 population segregating for 2 RFLPs can be used to map RFLPs relative to each other (see Fig 2 for example of mapping data between CUT1 + BEAU). 3) As illustrated in the map above, AG (a gene controlling flower development, which has been cloned – DNA probe available) is more tightly linked to CUT1 than BEAU. Show the results of an experiment that demonstrates this, building on the data in Fig. 2) [i.e. using the same DNA blot – you need to invent Ler and Col RFLP phenotypes for Ag and probe the same blot shown in Fig. 2]. 4) As illustrated in the map above, CER6 (also cloned; DNA probe available) is unlinked to CUT1 (on a different arm of the same chromosome). Show the results of an experiment that show this, building on the data in 2) [using the same DNA blot – you need to invent Ler and Col RFLP phenotypes for CER6]. 5) GL2 is the glaborous2 gene, identified only by leaf hair phenotype. The gene has not been cloned. G12 mutants are recessive and are easily distinguished from wild-type GL2 because leaves lack trichomes (leaf hairs) and thus appear shiny (glaborous) instead of fuzzy. To map GL2 relative to CUT1, a mapping population was set up as follows: gl2 gl2 X (Ler) ↓ gl2 GL2 X ↓ ¼ glaborous ¾ wild-type GL2 GL2 (Col) selfed P (phenotype) F2 population 100 individuals F1 Out of a population of 100 F2 plants, 25 were selected with the g12 phenotype. Show the results of an experiment using these 25 individuals that shows linkage of the GL2 gene to the CUT1 gene, with a map distance of 8 map units. [you will need to prepare a new DNA blot for this] D:\533564115.doc











